METRIC PREFIXES and SI UNITS Overview
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Basic Statistics Range, Mean, Median, Mode, Variance, Standard Deviation
Kirkup Chapter 1 Lecture Notes SEF Workshop presentation = Science Fair Data Analysis_Sohl.pptx EDA = Exploratory Data Analysis Aim of an Experiment = Experiments should have a goal. Make note of interesting issues for later. Be careful not to get side-tracked. Designing an Experiment = Put effort where it makes sense, do preliminary fractional error analysis to determine where you need to improve. Units analysis and equation checking Units website Writing down values: scientific notation and SI prefixes http://en.wikipedia.org/wiki/Metric_prefix Significant Figures Histograms in Excel Bin size – reasonable round numbers 15-16 not 14.8-16.1 Bin size – reasonable quantity: Rule of Thumb = # bins = √ where n is the number of data points. Basic Statistics range, mean, median, mode, variance, standard deviation Population, Sample Cool trick for small sample sizes. Estimate this works for n<12 or so. √ Random Error vs. Systematic Error Metric prefixes m n [n 1] Prefix Symbol 1000 10 Decimal Short scale Long scale Since 8 24 yotta Y 1000 10 1000000000000000000000000septillion quadrillion 1991 7 21 zetta Z 1000 10 1000000000000000000000sextillion trilliard 1991 6 18 exa E 1000 10 1000000000000000000quintillion trillion 1975 5 15 peta P 1000 10 1000000000000000quadrillion billiard 1975 4 12 tera T 1000 10 1000000000000trillion billion 1960 3 9 giga G 1000 10 1000000000billion milliard 1960 2 6 mega M 1000 10 1000000 million 1960 1 3 kilo k 1000 10 1000 thousand 1795 2/3 2 hecto h 1000 10 100 hundred 1795 1/3 1 deca da 1000 10 10 ten 1795 0 0 1000 -

Picosecond Multi-Hole Transfer and Microsecond Charge- Separated States at the Perovskite Nanocrystal/Tetracene Interface
Electronic Supplementary Material (ESI) for Chemical Science. This journal is © The Royal Society of Chemistry 2019 Electronic Supplementary Information for: Picosecond Multi-Hole Transfer and Microsecond Charge- Separated States at the Perovskite Nanocrystal/Tetracene Interface Xiao Luo, a,† Guijie Liang, ab,† Junhui Wang,a Xue Liu,a and Kaifeng Wua* a State Key Laboratory of Molecular Reaction Dynamics, Dynamics Research Center for Energy and Environmental Materials, and Collaborative Innovation Center of Chemistry for Energy Materials (iChEM), Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian, Liaoning 116023, China b Hubei Key Laboratory of Low Dimensional Optoelectronic Materials and Devices, Hubei University of Arts and Science, Xiangyang, Hubei 441053, China * Corresponding Author: [email protected] † Xiao Luo and Guijie Liang contributed equally to this work. Content list: Figures S1-S9 Sample preparations TA experiment set-ups Estimation of the number of attached TCA molecules per NC Estimation of FRET rate Estimation of average exciton number at each excitation density Estimation of exciton dissociation number S1 a 100 b long edge c short edge 70 7.90.1 nm 9.60.1 nm 60 80 50 s s 60 t t n 40 n u u o o C 30 C 40 20 20 10 0 0 4 6 8 10 12 14 16 18 2 4 6 8 10 12 Size (nm) Size (nm) Figure S1. (a) A representative transmission electron microscopy (TEM) image of CsPbBrxCl3-x perovskite NCs used in this study. Inset: high-resolution TEM. (b,c) Length distribution histograms of the NCs along the long (b) and short (c) edges. -

Time and Frequency Users' Manual
,>'.)*• r>rJfl HKra mitt* >\ « i If I * I IT I . Ip I * .aference nbs Publi- cations / % ^m \ NBS TECHNICAL NOTE 695 U.S. DEPARTMENT OF COMMERCE/National Bureau of Standards Time and Frequency Users' Manual 100 .U5753 No. 695 1977 NATIONAL BUREAU OF STANDARDS 1 The National Bureau of Standards was established by an act of Congress March 3, 1901. The Bureau's overall goal is to strengthen and advance the Nation's science and technology and facilitate their effective application for public benefit To this end, the Bureau conducts research and provides: (1) a basis for the Nation's physical measurement system, (2) scientific and technological services for industry and government, a technical (3) basis for equity in trade, and (4) technical services to pro- mote public safety. The Bureau consists of the Institute for Basic Standards, the Institute for Materials Research the Institute for Applied Technology, the Institute for Computer Sciences and Technology, the Office for Information Programs, and the Office of Experimental Technology Incentives Program. THE INSTITUTE FOR BASIC STANDARDS provides the central basis within the United States of a complete and consist- ent system of physical measurement; coordinates that system with measurement systems of other nations; and furnishes essen- tial services leading to accurate and uniform physical measurements throughout the Nation's scientific community, industry, and commerce. The Institute consists of the Office of Measurement Services, and the following center and divisions: Applied Mathematics -

Inflationary Big Bang Cosmology and the New Cosmic Background Radiation Findings
Inflationary Big Bang Cosmology and the New Cosmic Background Radiation Findings By Richard M. Todaro American Physical Society June 2001 With special thanks to Dr. Paul L. Richards, Professor of Physics at the University of California, Berkeley for his kind and patient telephone tutorial on Big Bang cosmology. A trio of new findings lends strong support to a powerful idea called “inflation” that explains many of the observed characteristics of our universe. The findings relate to the slight variations in the faint microwave energy that permeates the cosmos. The size and distribution of these variations agree well with the theory of inflation, which holds that the entire universe underwent an incredibly brief period of mind-bogglingly vast expansion (a hyper-charged Big Bang) before slowing down to the slower rate of expansion observed today. Combining the new findings about the cosmic microwave background radiation with inflation, cosmologists (the scientists who study the origin of the cosmos) believe they also have a compelling explanation for why stars and galaxies have their irregular distribution across the observable universe. Some cosmologists now believe that not only is the irregular distribution of “ordinary matter” related to the nature of these variations in the cosmic background radiation, but through inflation, both are related to the conditions within that incredibly dense and hot universe at the instant of its creation. (Such ordinary matter includes the elements that make up our Sun and our Earth and, ultimately, human beings.) Cosmologists have long believed that every bit of matter in the entire observable Universe – all the planets, stars, and galaxies, which we can see through powerful telescopes, as well as all the things we can’t see directly but know must be there due to the effects they produce – was once compressed together into a very dense and very hot arbitrarily small volume. -

Mathematics 3-4
Mathematics 3-4 Mathematics Department Phillips Exeter Academy Exeter, NH August 2021 To the Student Contents: Members of the PEA Mathematics Department have written the material in this book. As you work through it, you will discover that algebra, geometry, and trigonometry have been integrated into a mathematical whole. There is no Chapter 5, nor is there a section on tangents to circles. The curriculum is problem-centered, rather than topic-centered. Techniques and theorems will become apparent as you work through the problems, and you will need to keep appropriate notes for your records | there are no boxes containing important theorems. There is no index as such, but the reference section that starts on page 103 should help you recall the meanings of key words that are defined in the problems (where they usually appear italicized). Problem solving: Approach each problem as an exploration. Reading each question care- fully is essential, especially since definitions, highlighted in italics, are routinely inserted into the problem texts. It is important to make accurate diagrams. Here are a few useful strategies to keep in mind: create an easier problem, use the guess-and-check technique as a starting point, work backwards, recall work on a similar problem. It is important that you work on each problem when assigned, since the questions you may have about a problem will likely motivate class discussion the next day. Problem solving requires persistence as much as it requires ingenuity. When you get stuck, or solve a problem incorrectly, back up and start over. Keep in mind that you're probably not the only one who is stuck, and that may even include your teacher. -

Powers of 10 & Scientific Notation
Physics 151 Powers of 10 & Scientific Notation In all of the physical sciences, we encounter many numbers that are so large or small that it would be exceedingly cumbersome to write them with dozens of trailing or leading zeroes. Since our number system is “base-10” (based on powers of 10), it is far more convenient to write very large and very small numbers in a special exponential notation called scientific notation. In scientific notation, a number is rewritten as a simple decimal multiplied by 10 raised to some power, n, like this: x.xxxx... × 10n Powers of Ten Remember that the powers of 10 are as follows: 0 10 = 1 1 –1 1 10 = 10 10 = 0.1 = 10 2 –2 1 10 = 100 10 = 0.01 = 100 3 –3 1 10 = 1000 10 = 0.001 = 1000 4 –4 1 10 = 10,000 10 = 0.0001 = 10,000 ...and so forth. There are some important powers that we use often: 103 = 1000 = one thousand 10–3 = 0.001 = one thousandth 106 = 1,000,000 = one million 10–6 = 0.000 001 = one millionth 109 = 1,000,000,000 = one billion 10–9 = 0.000 000 001 = one billionth 1012 = 1,000,000,000,000 = one trillion 10–12 = 0.000 000 000 001 = one trillionth In the left-hand columns above, where n is positive, note that n is simply the same as the number of zeroes in the full written-out form of the number! However, in the right-hand columns where n is negative, note that |n| is one greater than the number of place-holding zeroes. -
Units and Conversions the Metric System Originates Back to the 1700S in France
Numeracy Introduction to Units and Conversions The metric system originates back to the 1700s in France. It is known as a decimal system because conversions between units are based on powers of ten. This is quite different to the Imperial system of units where every conversion has a unique value. A physical quantity is an attribute or property of a substance that can be expressed in a mathematical equation. A quantity, for example the amount of mass of a substance, is made up of a value and a unit. If a person has a mass of 72kg: the quantity being measured is Mass, the value of the measurement is 72 and the unit of measure is kilograms (kg). Another quantity is length (distance), for example the length of a piece of timber is 3.15m: the quantity being measured is length, the value of the measurement is 3.15 and the unit of measure is metres (m). A unit of measurement refers to a particular physical quantity. A metre describes length, a kilogram describes mass, a second describes time etc. A unit is defined and adopted by convention, in other words, everyone agrees that the unit will be a particular quantity. Historically, the metre was defined by the French Academy of Sciences as the length between two marks on a platinum-iridium bar at 0°C, which was designed to represent one ten-millionth of the distance from the Equator to the North Pole through Paris. In 1983, the metre was redefined as the distance travelled by light 1 in free space in of a second. -

Radiowave Propagation 1 Introduction Wireless Communication Relies on Electromagnetic Waves to Carry Information from One Point to Another
Lecture 2 Radiowave Propagation 1 Introduction Wireless communication relies on electromagnetic waves to carry information from one point to another. This propagation forms our physical channel. It is essential that we understand it if we are going to be able to understand and design wireless systems. This is a complex subject and will be our focus for several lectures. We will begin by considering the case of free-space propagation. First, however, we need to learn/review the dBm logarithmic power scale, as we will use this extensively. 2 Logarithmic Power Scales Power levels in radio systems can have very large dynamic ranges. For example, a CB (“citizen’s band”) radio might transmit 4W and receive as little as 10 fW ( 10−14 W ). That’s a dynamic range of more than 14 orders of magnitude. Instead of having to use factors like 1014 , it is usually more convenient to work on a logarithmic scale. Also, most of the systems we will consider can be treated as a cascade of blocks in which each block adds a gain factor. Since logarithms turn factors into sums, working on a log scale often simplifies analysis – you add instead of multiply. A dimensionless number A can be represented on a decibel scale as A =10log A dB (1) A 10 A=10 dB / If P is a power level then it has dimensions. Therefore, we cannot directly convert it to a dB scale. But we can get a dimensionless number by taking the ratio of P to some reference power. The most commonly used reference in RF telecom is one milliwatt, and we write dBm, meaning dB relative to a milliwatt. -
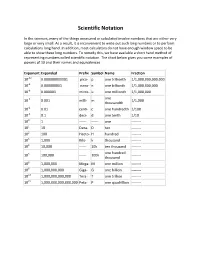
Scientific Notation
Scientific Notation In the sciences, many of the things measured or calculated involve numbers that are either very large or very small. As a result, it is inconvenient to write out such long numbers or to perform calculations long hand. In addition, most calculators do not have enough window space to be able to show these long numbers. To remedy this, we have available a short hand method of representing numbers called scientific notation. The chart below gives you some examples of powers of 10 and their names and equivalences. Exponent Expanded Prefix Symbol Name Fraction 10-12 0.000000000001 pico- p one trillionth 1/1,000,000,000,000 10-9 0.000000001 nano- n one billionth 1/1,000,000,000 10-6 0.000001 micro- u one millionth 1/1,000,000 one 10-3 0.001 milli- m 1/1,000 thousandth 10-2 0.01 centi- c one hundredth 1/100 10-1 0.1 deci- d one tenth 1/10 100 1 ------ ------ one -------- 101 10 Deca- D ten -------- 102 100 Hecto- H hundred -------- 103 1,000 Kilo- k thousand -------- 104 10,000 ------ 10k ten thousand -------- one hundred 105 100,000 ------ 100k -------- thousand 106 1,000,000 Mega- M one million -------- 109 1,000,000,000 Giga- G one billion -------- 1012 1,000,000,000,000 Tera- T one trillion -------- 1015 1,000,000,000,000,000 Peta- P one quadrillion -------- Powers of 10 and Place Value.......... Multiplying by 10, 100, or 1000 in the following problems just means to add the number of zeroes to the number being multiplied. -

The Planck Time Unit (5.39 × 10 S) Is the Time It Takes Light to Travel One
GNSS Applications (Precise Timing 1) In addition to longitude, latitude, and altitude, the GNSS systems provide a critical fourth dimension – time. Each GNSS satellite carries two atomic clocks that contribute very precise time data to the GNSS signals. GNSS receivers decode these signals, effectively synchronising each receiver to the atomic clocks. This enables users to determine the time to within 100 billionths of a second, without the cost of owning and operating atomic clocks. In June 2012, at the US Track & Field Trials for the London Olympics, Allyson Felix and Jeneba crossed the finishing line of the 100m both with a time of 11.068 seconds. Even using a high speed camera shooting 3,000 frames per second (fps), the pair could not be separated. 3,0000 fps is ~3 times faster than the time it takes the average human to blink an eye; however, even this is slow when compared to GNSS time, which can measure time in nanoseconds, that is one billionth of a second To put nanoseconds into perspective, if a nanosecond was scaled up to one second, at this scale a second would be 31 years long. Other units of time include: Decisecond 0.1s Centisecond 0.01s Millisecond 0.001s Microsecond 10-6s -9 Nanosecond 10 s Picosecond 10-12s Femtosecond 10-15s The Planck Time Unit (5.39 × 10-44s) is Attosecond 10-18s (the shortest the time it takes light to travel one -35 length of time currently measurable) Planck length (1.616199 × 10 m) GNSS Applications (Financial Markets) ‘Time is money’ so the saying goes and with the advent of GPS (Global Positioning System) becoming publicly available in 1983, this has become increasingly true in a truly surprising way. -

Rare-Earth Elements
Rare-Earth Elements Chapter O of Critical Mineral Resources of the United States—Economic and Environmental Geology and Prospects for Future Supply Professional Paper 1802–O U.S. Department of the Interior U.S. Geological Survey Periodic Table of Elements 1A 8A 1 2 hydrogen helium 1.008 2A 3A 4A 5A 6A 7A 4.003 3 4 5 6 7 8 9 10 lithium beryllium boron carbon nitrogen oxygen fluorine neon 6.94 9.012 10.81 12.01 14.01 16.00 19.00 20.18 11 12 13 14 15 16 17 18 sodium magnesium aluminum silicon phosphorus sulfur chlorine argon 22.99 24.31 3B 4B 5B 6B 7B 8B 11B 12B 26.98 28.09 30.97 32.06 35.45 39.95 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 potassium calcium scandium titanium vanadium chromium manganese iron cobalt nickel copper zinc gallium germanium arsenic selenium bromine krypton 39.10 40.08 44.96 47.88 50.94 52.00 54.94 55.85 58.93 58.69 63.55 65.39 69.72 72.64 74.92 78.96 79.90 83.79 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 rubidium strontium yttrium zirconium niobium molybdenum technetium ruthenium rhodium palladium silver cadmium indium tin antimony tellurium iodine xenon 85.47 87.62 88.91 91.22 92.91 95.96 (98) 101.1 102.9 106.4 107.9 112.4 114.8 118.7 121.8 127.6 126.9 131.3 55 56 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 cesium barium hafnium tantalum tungsten rhenium osmium iridium platinum gold mercury thallium lead bismuth polonium astatine radon 132.9 137.3 178.5 180.9 183.9 186.2 190.2 192.2 195.1 197.0 200.5 204.4 207.2 209.0 (209) (210) (222) 87 88 104 105 106 107 108 109 110 111 112 113 114 115 -

Dynamically Driven Protein Allostery Exhibits Disparate Responses for Fast and Slow Motions
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Biophysical Journal Volume 108 June 2015 2771–2774 2771 Biophysical Letter Dynamically Driven Protein Allostery Exhibits Disparate Responses for Fast and Slow Motions Jingjing Guo1,2 and Huan-Xiang Zhou1,* 1Department of Physics and Institute of Molecular Biophysics, Florida State University, Tallahassee, Florida; and 2School of Pharmacy, State Key Laboratory of Applied Organic Chemistry and Department of Chemistry, Lanzhou University, Lanzhou, China ABSTRACT There is considerable interest in the dynamic aspect of allosteric action, and in a growing list of proteins allostery has been characterized as being mediated predominantly by a change in dynamics, not a transition in conformation. For consid- ering conformational dynamics, a protein molecule can be simplified into a number of relatively rigid microdomains connected by joints, corresponding to, e.g., communities and edges from a community network analysis. Binding of an allosteric activator strengthens intermicrodomain coupling, thereby quenching fast (e.g., picosecond to nanosecond) local motions but initiating slow (e.g., microsecond to millisecond), cross-microdomain correlated motions that are potentially of functional importance. This scenario explains allosteric effects observed in many unrelated proteins. Received for publication 20 March 2015 and in final form 24 April 2015. *Correspondence: [email protected] Allostery, whereby ligand binding at a distal site affects Our simple model, inspired by community analysis on binding affinity or catalytic activity at the active site, is Pin1 (12), represents three main microdomains of Pin1 as traditionally considered as mediated through conforma- springs for describing local motions (Fig.