Atlas Journal
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Wnt/Β-Catenin Signaling Pathway Induces Autophagy
Yun et al. Cell Death and Disease (2020) 11:771 https://doi.org/10.1038/s41419-020-02988-8 Cell Death & Disease ARTICLE Open Access Wnt/β-catenin signaling pathway induces autophagy-mediated temozolomide-resistance in human glioblastoma Eun-Jin Yun 1,SangwooKim2,Jer-TsongHsieh3,4 and Seung Tae Baek 5,6 Abstract Temozolomide (TMZ) is widely used for treating glioblastoma multiforme (GBM), however, the treatment of such brain tumors remains a challenge due to the development of resistance. Increasing studies have found that TMZ treatment could induce autophagy that may link to therapeutic resistance in GBM, but, the precise mechanisms are not fully understood. Understanding the molecular mechanisms underlying the response of GBM to chemotherapy is paramount for developing improved cancer therapeutics. In this study, we demonstrated that the loss of DOC-2/DAB2 interacting protein (DAB2IP) is responsible for TMZ-resistance in GBM through ATG9B. DAB2IP sensitized GBM to TMZ and suppressed TMZ-induced autophagy by negatively regulating ATG9B expression. A higher level of ATG9B expression was associated with GBM compared to low-grade glioma. The knockdown of ATG9B expression in GBM cells suppressed TMZ-induced autophagy as well as TMZ-resistance. Furthermore, we showed that DAB2IP negatively regulated ATG9B expression by blocking the Wnt/β-catenin pathway. To enhance the benefit of TMZ and avoid therapeutic resistance, effective combination strategies were tested using a small molecule inhibitor blocking the Wnt/ β-catenin pathway in addition to TMZ. The combination treatment synergistically enhanced the efficacy of TMZ in GBM cells. In conclusion, the present study identified the mechanisms of TMZ-resistance of GBM mediated by DAB2IP 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; and ATG9B which provides insight into a potential strategy to overcome TMZ chemo-resistance. -

Rabbit Anti-DAB2IP Rabbit Anti-DAB2IP
Qty: 100 μg/400 μL Rabbit anti-DAB2IP Catalog No. 487300 Lot No. Rabbit anti-DAB2IP FORM This polyclonal antibody is supplied as a 400 µL aliquot at a concentration of 0.25 mg/mL in phosphate buffered saline (pH 7.4) containing 0.1% sodium azide. This antibody is epitope-affinity purified from rabbit antiserum. PAD: ZMD.689 IMMUNOGEN Synthetic peptide derived from the C-terminal region of the human DAB2IP protein (Accession# NP_619723), which is identical to mouse and rat sequence. SPECIFICITY This antibody is specific for the DAB2IP (DAB2 interacting protein, AIP1, DIP1/2) protein. On Western blots, it identifies the target band at ~110 kDa. REACTIVITY Reactivity has been confirmed with human DU145, SK-N-MC and rat B49 cell lysates. Based on amino acid sequence homology, reactivity with mouse is expected. Sample Western Immuno- Immuno- Blotting precipitation cytochemistry Human +++ 0 ND Mouse ND ND ND Rat +++ 0 ND (Excellent +++, Good++, Poor +, No reactivity 0, Not applicable N/A, Not Determined ND) USAGE Working concentrations for specific applications should be determined by the investigator. Appropriate concentrations will be affected by several factors, including secondary antibody affinity, antigen concentration, sensitivity of detection method, temperature and length of incubations, etc. The suitability of this antibody for applications other than those listed below has not been determined. The following concentration ranges are recommended starting points for this product. Western Blotting: 1-3 μg/mL STORAGE Store at 2-8°C for up to one month. Store at –20°C for long-term storage. Avoid repeated freezing and thawing. (cont’d) www.invitrogen.com Invitrogen Corporation • 542 Flynn Rd • Camarillo • CA 93012 • Tel: 800.955.6288 • E-mail: [email protected] PI487300 (Rev 10/08) DCC-08-1089 Important Licensing Information - These products may be covered by one or more Limited Use Label Licenses (see the Invitrogen Catalog or our website, www.invitrogen.com). -

Disabled Homolog 2 Interactive Protein Functions As a Tumor Suppressor in Osteosarcoma Cells
ONCOLOGY LETTERS 16: 703-712, 2018 Disabled homolog 2 interactive protein functions as a tumor suppressor in osteosarcoma cells JIANAN HE1,5, SHUAI HUANG2, ZHENHUA LIN3, JIQIN ZHANG4, JIALIN SU4, WEIDONG JI4 and XINGMO LIU1 1Department of Orthopaedic Surgery, The Sixth Affiliated Hospital of Sun Yat‑sun University, Guangzhou, Guangdong 510655; 2Department of Orthopaedic Surgery, The First Affiliated Hospital of Sun Yat‑sen University, Guangzhou, Guangdong 510080; 3Department of Orthopaedic Surgery, The People's Hospital of Guangxi Zhuang Autonomous Region, Nanning, Guangxi 530021; 4Center for Translational Medicine, The First Affiliated Hospital of Sun Yat‑sen University, Guangzhou, Guangdong 510080; 5Department of Interventional Radiology, The Fifth Affiliated Hospital of Sun Yat-sun University, Zhuhai, Guangdong 519000, P.R. China Received April 2, 2016; Accepted June 16, 2017 DOI: 10.3892/ol.2018.8776 Abstract. The disabled homolog 2 interactive protein osteosarcoma will develop distant metastasis (2). Owing to the (DAB2IP) gene is a member of the family of Ras GTPases development of multidisciplinary therapy, the mortality rate and functions as a tumor suppressor in many types of carci- for patients with osteosarcoma has been decreasing in recent noma; however, its function in osteosarcoma remains unclear. years. Nevertheless, patients continue to exhibit a high risk The aim of the present study was to determine the function of of metastasis and/or recurrence (1,2). Previous studies have DAB2IP in osteosarcoma and normal bone cells in vitro. The indicated that several genetic aberrations are associated with expression of DAB2IP protein was assessed in osteoblast and tumor initiation and osteosarcoma progression (3,4). Thus, osteosarcoma cell lines by western blot analysis. -

Methylome and Transcriptome Maps of Human Visceral and Subcutaneous
www.nature.com/scientificreports OPEN Methylome and transcriptome maps of human visceral and subcutaneous adipocytes reveal Received: 9 April 2019 Accepted: 11 June 2019 key epigenetic diferences at Published: xx xx xxxx developmental genes Stephen T. Bradford1,2,3, Shalima S. Nair1,3, Aaron L. Statham1, Susan J. van Dijk2, Timothy J. Peters 1,3,4, Firoz Anwar 2, Hugh J. French 1, Julius Z. H. von Martels1, Brodie Sutclife2, Madhavi P. Maddugoda1, Michelle Peranec1, Hilal Varinli1,2,5, Rosanna Arnoldy1, Michael Buckley1,4, Jason P. Ross2, Elena Zotenko1,3, Jenny Z. Song1, Clare Stirzaker1,3, Denis C. Bauer2, Wenjia Qu1, Michael M. Swarbrick6, Helen L. Lutgers1,7, Reginald V. Lord8, Katherine Samaras9,10, Peter L. Molloy 2 & Susan J. Clark 1,3 Adipocytes support key metabolic and endocrine functions of adipose tissue. Lipid is stored in two major classes of depots, namely visceral adipose (VA) and subcutaneous adipose (SA) depots. Increased visceral adiposity is associated with adverse health outcomes, whereas the impact of SA tissue is relatively metabolically benign. The precise molecular features associated with the functional diferences between the adipose depots are still not well understood. Here, we characterised transcriptomes and methylomes of isolated adipocytes from matched SA and VA tissues of individuals with normal BMI to identify epigenetic diferences and their contribution to cell type and depot-specifc function. We found that DNA methylomes were notably distinct between diferent adipocyte depots and were associated with diferential gene expression within pathways fundamental to adipocyte function. Most striking diferential methylation was found at transcription factor and developmental genes. Our fndings highlight the importance of developmental origins in the function of diferent fat depots. -

Negative Regulation of DAB2IP by Akt and Scffbw7 Pathways
Negative regulation of DAB2IP by Akt and SCFFbw7 pathways The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Dai, Xiangping, Brian J. North, and Hiroyuki Inuzuka. 2014. “Negative regulation of DAB2IP by Akt and SCFFbw7 pathways.” Oncotarget 5 (10): 3307-3315. Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:12717538 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA www.impactjournals.com/oncotarget/ Oncotarget, Vol. 5, No. 10 Negative regulation of DAB2IP by Akt and SCFFbw7 pathways Xiangping Dai1,*, Brian J. North1,* and Hiroyuki Inuzuka1 1 Department of Pathology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA. * These authors contributed equally to this work. Correspondence to: Brian North, email: [email protected] Correspondence to: Hiroyuki Inuzuka, email: [email protected] Keywords: DAB2IP, Akt, Fbw7, degradation, cancer, phosphorylation, ubiquitination. Received: April 16, 2014 Accepted: April 30, 2014 Published: May 1, 2014 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT: Deletion of ovarian carcinoma 2/disabled homolog 2 (DOC-2/DAB2) interacting protein (DAB2IP), is a tumor suppressor that serves as a scaffold protein involved in coordinately regulating cell proliferation, survival and apoptotic pathways. -

(P -Value<0.05, Fold Change≥1.4), 4 Vs. 0 Gy Irradiation
Table S1: Significant differentially expressed genes (P -Value<0.05, Fold Change≥1.4), 4 vs. 0 Gy irradiation Genbank Fold Change P -Value Gene Symbol Description Accession Q9F8M7_CARHY (Q9F8M7) DTDP-glucose 4,6-dehydratase (Fragment), partial (9%) 6.70 0.017399678 THC2699065 [THC2719287] 5.53 0.003379195 BC013657 BC013657 Homo sapiens cDNA clone IMAGE:4152983, partial cds. [BC013657] 5.10 0.024641735 THC2750781 Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). 4.07 0.04353262 DNAH5 [Source:Uniprot/SWISSPROT;Acc:Q8TE73] [ENST00000382416] 3.81 0.002855909 NM_145263 SPATA18 Homo sapiens spermatogenesis associated 18 homolog (rat) (SPATA18), mRNA [NM_145263] AA418814 zw01a02.s1 Soares_NhHMPu_S1 Homo sapiens cDNA clone IMAGE:767978 3', 3.69 0.03203913 AA418814 AA418814 mRNA sequence [AA418814] AL356953 leucine-rich repeat-containing G protein-coupled receptor 6 {Homo sapiens} (exp=0; 3.63 0.0277936 THC2705989 wgp=1; cg=0), partial (4%) [THC2752981] AA484677 ne64a07.s1 NCI_CGAP_Alv1 Homo sapiens cDNA clone IMAGE:909012, mRNA 3.63 0.027098073 AA484677 AA484677 sequence [AA484677] oe06h09.s1 NCI_CGAP_Ov2 Homo sapiens cDNA clone IMAGE:1385153, mRNA sequence 3.48 0.04468495 AA837799 AA837799 [AA837799] Homo sapiens hypothetical protein LOC340109, mRNA (cDNA clone IMAGE:5578073), partial 3.27 0.031178378 BC039509 LOC643401 cds. [BC039509] Homo sapiens Fas (TNF receptor superfamily, member 6) (FAS), transcript variant 1, mRNA 3.24 0.022156298 NM_000043 FAS [NM_000043] 3.20 0.021043295 A_32_P125056 BF803942 CM2-CI0135-021100-477-g08 CI0135 Homo sapiens cDNA, mRNA sequence 3.04 0.043389246 BF803942 BF803942 [BF803942] 3.03 0.002430239 NM_015920 RPS27L Homo sapiens ribosomal protein S27-like (RPS27L), mRNA [NM_015920] Homo sapiens tumor necrosis factor receptor superfamily, member 10c, decoy without an 2.98 0.021202829 NM_003841 TNFRSF10C intracellular domain (TNFRSF10C), mRNA [NM_003841] 2.97 0.03243901 AB002384 C6orf32 Homo sapiens mRNA for KIAA0386 gene, partial cds. -

Human Lectins, Their Carbohydrate Affinities and Where to Find Them
biomolecules Review Human Lectins, Their Carbohydrate Affinities and Where to Review HumanFind Them Lectins, Their Carbohydrate Affinities and Where to FindCláudia ThemD. Raposo 1,*, André B. Canelas 2 and M. Teresa Barros 1 1, 2 1 Cláudia D. Raposo * , Andr1 é LAQVB. Canelas‐Requimte,and Department M. Teresa of Chemistry, Barros NOVA School of Science and Technology, Universidade NOVA de Lisboa, 2829‐516 Caparica, Portugal; [email protected] 12 GlanbiaLAQV-Requimte,‐AgriChemWhey, Department Lisheen of Chemistry, Mine, Killoran, NOVA Moyne, School E41 of ScienceR622 Co. and Tipperary, Technology, Ireland; canelas‐ [email protected] NOVA de Lisboa, 2829-516 Caparica, Portugal; [email protected] 2* Correspondence:Glanbia-AgriChemWhey, [email protected]; Lisheen Mine, Tel.: Killoran, +351‐212948550 Moyne, E41 R622 Tipperary, Ireland; [email protected] * Correspondence: [email protected]; Tel.: +351-212948550 Abstract: Lectins are a class of proteins responsible for several biological roles such as cell‐cell in‐ Abstract:teractions,Lectins signaling are pathways, a class of and proteins several responsible innate immune for several responses biological against roles pathogens. such as Since cell-cell lec‐ interactions,tins are able signalingto bind to pathways, carbohydrates, and several they can innate be a immuneviable target responses for targeted against drug pathogens. delivery Since sys‐ lectinstems. In are fact, able several to bind lectins to carbohydrates, were approved they by canFood be and a viable Drug targetAdministration for targeted for drugthat purpose. delivery systems.Information In fact, about several specific lectins carbohydrate were approved recognition by Food by andlectin Drug receptors Administration was gathered for that herein, purpose. plus Informationthe specific organs about specific where those carbohydrate lectins can recognition be found by within lectin the receptors human was body. -
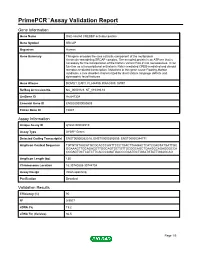
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name Snf2-related CREBBP activator protein Gene Symbol SRCAP Organism Human Gene Summary This gene encodes the core catalytic component of the multiprotein chromatin-remodeling SRCAP complex. The encoded protein is an ATPase that is necessary for the incorporation of the histone variant H2A.Z into nucleosomes. It can function as a transcriptional activator in Notch-mediated CREB-mediated and steroid receptor-mediated transcription. Mutations in this gene cause Floating-Harbor syndrome a rare disorder characterized by short stature language deficits and dysmorphic facial features. Gene Aliases DOMO1, EAF1, FLJ44499, KIAA0309, SWR1 RefSeq Accession No. NC_000016.9, NT_010393.16 UniGene ID Hs.647334 Ensembl Gene ID ENSG00000080603 Entrez Gene ID 10847 Assay Information Unique Assay ID qHsaCID0006510 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000262518, ENST00000395059, ENST00000344771 Amplicon Context Sequence TGTGTGTAACATGCGCACCCAGTTCCCTGACTTAAGACTCATCCAGTATGATTGC GGAAAGTTGCAGACGTTGGCAGTGCTGTTGCGGCAGCTCAAGGCAGAGGGCCA CCGAGTGCTCATCTTCACCCAGATGACCCGAATGCTGGATGTATTGGAGCAG Amplicon Length (bp) 130 Chromosome Location 16:30740838-30744704 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 90 R2 0.9977 cDNA Cq 19.2 cDNA Tm (Celsius) 86.5 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report SRCAP, Human -

In Silico Characterization of Class II Plant Defensins from Arabidopsis
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.27.065185; this version posted September 7, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. In silico Characterization of Class II Plant Defensins from Arabidopsis thaliana Laura S.M. Costa1,2, Állan S. Pires1, Neila B. Damaceno1, Pietra O. Rigueiras1, Mariana R. Maximiano1, Octavio L. Franco1,2,3, William F. Porto3,4* 1 Centro de Análises Proteômicas e Bioquímicas. Programa de Pós-Graduação em Ciências Genômicas e Biotecnologia, Universidade Católica de Brasília, Brasília-DF, Brazil. 2 Departamento de Biologia, Programa de Pós-Graduação em Genética e Biotecnologia, Universidade Federal de Juiz de Fora, Campus Universitário, Juiz de Fora-MG, Brazil. 3 S-Inova Biotech, Pós-Graduação em Biotecnologia, Universidade Católica Dom Bosco, Campo Grande-MS, Brazil. 4 Porto Reports, Brasília-DF, Brazil – www.portoreports.com *Corresponding author: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.04.27.065185; this version posted September 7, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract Defensins comprise a polyphyletic group of multifunctional defense peptides. Cis- defensins, also known as cysteine stabilized αβ (CSαβ) defensins, are one of the most ancient defense peptide families. -

Diagnostic Interpretation of Genetic Studies in Patients with Primary
AAAAI Work Group Report Diagnostic interpretation of genetic studies in patients with primary immunodeficiency diseases: A working group report of the Primary Immunodeficiency Diseases Committee of the American Academy of Allergy, Asthma & Immunology Ivan K. Chinn, MD,a,b Alice Y. Chan, MD, PhD,c Karin Chen, MD,d Janet Chou, MD,e,f Morna J. Dorsey, MD, MMSc,c Joud Hajjar, MD, MS,a,b Artemio M. Jongco III, MPH, MD, PhD,g,h,i Michael D. Keller, MD,j Lisa J. Kobrynski, MD, MPH,k Attila Kumanovics, MD,l Monica G. Lawrence, MD,m Jennifer W. Leiding, MD,n,o,p Patricia L. Lugar, MD,q Jordan S. Orange, MD, PhD,r,s Kiran Patel, MD,k Craig D. Platt, MD, PhD,e,f Jennifer M. Puck, MD,c Nikita Raje, MD,t,u Neil Romberg, MD,v,w Maria A. Slack, MD,x,y Kathleen E. Sullivan, MD, PhD,v,w Teresa K. Tarrant, MD,z Troy R. Torgerson, MD, PhD,aa,bb and Jolan E. Walter, MD, PhDn,o,cc Houston, Tex; San Francisco, Calif; Salt Lake City, Utah; Boston, Mass; Great Neck and Rochester, NY; Washington, DC; Atlanta, Ga; Rochester, Minn; Charlottesville, Va; St Petersburg, Fla; Durham, NC; Kansas City, Mo; Philadelphia, Pa; and Seattle, Wash AAAAI Position Statements,Work Group Reports, and Systematic Reviews are not to be considered to reflect current AAAAI standards or policy after five years from the date of publication. The statement below is not to be construed as dictating an exclusive course of action nor is it intended to replace the medical judgment of healthcare professionals. -

LJELSR: a Strengthened Version of JELSR for Feature Selection and Clustering
Article LJELSR: A Strengthened Version of JELSR for Feature Selection and Clustering Sha-Sha Wu 1, Mi-Xiao Hou 1, Chun-Mei Feng 1,2 and Jin-Xing Liu 1,* 1 School of Information Science and Engineering, Qufu Normal University, Rizhao 276826, China; [email protected] (S.-S.W.); [email protected] (M.-X.H.); [email protected] (C.-M.F.) 2 Bio-Computing Research Center, Harbin Institute of Technology, Shenzhen 518055, China * Correspondence: [email protected]; Tel.: +086-633-3981-241 Received: 4 December 2018; Accepted: 7 February 2019; Published: 18 February 2019 Abstract: Feature selection and sample clustering play an important role in bioinformatics. Traditional feature selection methods separate sparse regression and embedding learning. Later, to effectively identify the significant features of the genomic data, Joint Embedding Learning and Sparse Regression (JELSR) is proposed. However, since there are many redundancy and noise values in genomic data, the sparseness of this method is far from enough. In this paper, we propose a strengthened version of JELSR by adding the L1-norm constraint on the regularization term based on a previous model, and call it LJELSR, to further improve the sparseness of the method. Then, we provide a new iterative algorithm to obtain the convergence solution. The experimental results show that our method achieves a state-of-the-art level both in identifying differentially expressed genes and sample clustering on different genomic data compared to previous methods. Additionally, the selected differentially expressed genes may be of great value in medical research. Keywords: differentially expressed genes; feature selection; L1-norm; sample clustering; sparse constraint 1. -

Dynamics of Transcription Elongation Are Finely Tuned by Dozens of Regulatory Factors
bioRxiv preprint doi: https://doi.org/10.1101/2021.08.15.456358; this version posted August 15, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Dynamics of transcription elongation are finely tuned by dozens of regulatory factors Mary Couvillion1*, Kevin M. Harlen1*, Kate C. Lachance1*, Kristine L. Trotta1, Erin Smith1, Christian Brion1, Brendan M. Smalec1, L. Stirling Churchman1 1Blavatnik Institute, Department of Genetics, Harvard Medical School, Boston, Massachusetts, USA 02115 *these authors contributed equally 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.08.15.456358; this version posted August 15, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. ABSTRACT Understanding the complex network and dynamics that regulate transcription elongation requires the quantitative analysis of RNA polymerase II (Pol II) activity in a wide variety of regulatory environments. We performed native elongating transcript sequencing (NET-seq) in 41 strains of S. cerevisiae lacking known elongation regulators, including RNA processing factors, transcription elongation factors, chromatin modifiers, and remodelers. We found that the opposing effects of these factors balance transcription elongation dynamics. Different sets of factors tightly regulate Pol II progression across gene bodies so that Pol II density peaks at key points of RNA processing.