Randomization-Based Inference for Bernoulli Trial Experiments And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

3.2.3 Binomial Distribution
3.2.3 Binomial Distribution The binomial distribution is based on the idea of a Bernoulli trial. A Bernoulli trail is an experiment with two, and only two, possible outcomes. A random variable X has a Bernoulli(p) distribution if 8 > <1 with probability p X = > :0 with probability 1 − p, where 0 ≤ p ≤ 1. The value X = 1 is often termed a “success” and X = 0 is termed a “failure”. The mean and variance of a Bernoulli(p) random variable are easily seen to be EX = (1)(p) + (0)(1 − p) = p and VarX = (1 − p)2p + (0 − p)2(1 − p) = p(1 − p). In a sequence of n identical, independent Bernoulli trials, each with success probability p, define the random variables X1,...,Xn by 8 > <1 with probability p X = i > :0 with probability 1 − p. The random variable Xn Y = Xi i=1 has the binomial distribution and it the number of sucesses among n independent trials. The probability mass function of Y is µ ¶ ¡ ¢ n ¡ ¢ P Y = y = py 1 − p n−y. y For this distribution, t n EX = np, Var(X) = np(1 − p),MX (t) = [pe + (1 − p)] . 1 Theorem 3.2.2 (Binomial theorem) For any real numbers x and y and integer n ≥ 0, µ ¶ Xn n (x + y)n = xiyn−i. i i=0 If we take x = p and y = 1 − p, we get µ ¶ Xn n 1 = (p + (1 − p))n = pi(1 − p)n−i. i i=0 Example 3.2.2 (Dice probabilities) Suppose we are interested in finding the probability of obtaining at least one 6 in four rolls of a fair die. -

5 Stochastic Processes
5 Stochastic Processes Contents 5.1. The Bernoulli Process ...................p.3 5.2. The Poisson Process .................. p.15 1 2 Stochastic Processes Chap. 5 A stochastic process is a mathematical model of a probabilistic experiment that evolves in time and generates a sequence of numerical values. For example, a stochastic process can be used to model: (a) the sequence of daily prices of a stock; (b) the sequence of scores in a football game; (c) the sequence of failure times of a machine; (d) the sequence of hourly traffic loads at a node of a communication network; (e) the sequence of radar measurements of the position of an airplane. Each numerical value in the sequence is modeled by a random variable, so a stochastic process is simply a (finite or infinite) sequence of random variables and does not represent a major conceptual departure from our basic framework. We are still dealing with a single basic experiment that involves outcomes gov- erned by a probability law, and random variables that inherit their probabilistic † properties from that law. However, stochastic processes involve some change in emphasis over our earlier models. In particular: (a) We tend to focus on the dependencies in the sequence of values generated by the process. For example, how do future prices of a stock depend on past values? (b) We are often interested in long-term averages,involving the entire se- quence of generated values. For example, what is the fraction of time that a machine is idle? (c) We sometimes wish to characterize the likelihood or frequency of certain boundary events. -

1 Normal Distribution
1 Normal Distribution. 1.1 Introduction A Bernoulli trial is simple random experiment that ends in success or failure. A Bernoulli trial can be used to make a new random experiment by repeating the Bernoulli trial and recording the number of successes. Now repeating a Bernoulli trial a large number of times has an irritating side e¤ect. Suppose we take a tossed die and look for a 3 to come up, but we do this 6000 times. 1 This is a Bernoulli trial with a probability of success of 6 repeated 6000 times. What is the probability that we will see exactly 1000 success? This is de…nitely the most likely possible outcome, 1000 successes out of 6000 tries. But it is still very unlikely that an particular experiment like this will turn out so exactly. In fact, if 6000 tosses did produce exactly 1000 successes, that would be rather suspicious. The probability of exactly 1000 successes in 6000 tries almost does not need to be calculated. whatever the probability of this, it will be very close to zero. It is probably too small a probability to be of any practical use. It turns out that it is not all that bad, 0:014. Still this is small enough that it means that have even a chance of seeing it actually happen, we would need to repeat the full experiment as many as 100 times. All told, 600,000 tosses of a die. When we repeat a Bernoulli trial a large number of times, it is unlikely that we will be interested in a speci…c number of successes, and much more likely that we will be interested in the event that the number of successes lies within a range of possibilities. -

Bernoulli Random Forests: Closing the Gap Between Theoretical Consistency and Empirical Soundness
Proceedings of the Twenty-Fifth International Joint Conference on Artificial Intelligence (IJCAI-16) Bernoulli Random Forests: Closing the Gap between Theoretical Consistency and Empirical Soundness , , , ? Yisen Wang† ‡, Qingtao Tang† ‡, Shu-Tao Xia† ‡, Jia Wu , Xingquan Zhu ⇧ † Dept. of Computer Science and Technology, Tsinghua University, China ‡ Graduate School at Shenzhen, Tsinghua University, China ? Quantum Computation & Intelligent Systems Centre, University of Technology Sydney, Australia ⇧ Dept. of Computer & Electrical Engineering and Computer Science, Florida Atlantic University, USA wangys14, tqt15 @mails.tsinghua.edu.cn; [email protected]; [email protected]; [email protected] { } Traditional Bernoulli Trial Controlled Abstract Tree Node Splitting Tree Node Splitting Random forests are one type of the most effective ensemble learning methods. In spite of their sound Random Attribute Bagging Bernoulli Trial Controlled empirical performance, the study on their theoreti- Attribute Bagging cal properties has been left far behind. Recently, several random forests variants with nice theoreti- Random Structure/Estimation cal basis have been proposed, but they all suffer Random Bootstrap Sampling Points Splitting from poor empirical performance. In this paper, we (a) Breiman RF (b) BRF propose a Bernoulli random forests model (BRF), which intends to close the gap between the theoreti- Figure 1: Comparisons between Breiman RF (left panel) vs. cal consistency and the empirical soundness of ran- the proposed BRF (right panel). The tree node splitting of dom forests classification. Compared to Breiman’s Breiman RF is deterministic, so the final trees are highly data- original random forests, BRF makes two simplifi- dependent. Instead, BRF employs two Bernoulli distributions cations in tree construction by using two indepen- to control the tree construction. -

Probability Theory
Probability Theory Course Notes — Harvard University — 2011 C. McMullen March 29, 2021 Contents I TheSampleSpace ........................ 2 II Elements of Combinatorial Analysis . 5 III RandomWalks .......................... 15 IV CombinationsofEvents . 24 V ConditionalProbability . 29 VI The Binomial and Poisson Distributions . 37 VII NormalApproximation. 44 VIII Unlimited Sequences of Bernoulli Trials . 55 IX Random Variables and Expectation . 60 X LawofLargeNumbers...................... 68 XI Integral–Valued Variables. Generating Functions . 70 XIV RandomWalkandRuinProblems . 70 I The Exponential and the Uniform Density . 75 II Special Densities. Randomization . 94 These course notes accompany Feller, An Introduction to Probability Theory and Its Applications, Wiley, 1950. I The Sample Space Some sources and uses of randomness, and philosophical conundrums. 1. Flipped coin. 2. The interrupted game of chance (Fermat). 3. The last roll of the game in backgammon (splitting the stakes at Monte Carlo). 4. Large numbers: elections, gases, lottery. 5. True randomness? Quantum theory. 6. Randomness as a model (in reality only one thing happens). Paradox: what if a coin keeps coming up heads? 7. Statistics: testing a drug. When is an event good evidence rather than a random artifact? 8. Significance: among 1000 coins, if one comes up heads 10 times in a row, is it likely to be a 2-headed coin? Applications to economics, investment and hiring. 9. Randomness as a tool: graph theory; scheduling; internet routing. We begin with some previews. Coin flips. What are the chances of 10 heads in a row? The probability is 1/1024, less than 0.1%. Implicit assumptions: no biases and independence. 10 What are the chance of heads 5 out of ten times? ( 5 = 252, so 252/1024 = 25%). -
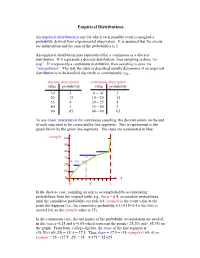
Discrete Distributions: Empirical, Bernoulli, Binomial, Poisson
Empirical Distributions An empirical distribution is one for which each possible event is assigned a probability derived from experimental observation. It is assumed that the events are independent and the sum of the probabilities is 1. An empirical distribution may represent either a continuous or a discrete distribution. If it represents a discrete distribution, then sampling is done “on step”. If it represents a continuous distribution, then sampling is done via “interpolation”. The way the table is described usually determines if an empirical distribution is to be handled discretely or continuously; e.g., discrete description continuous description value probability value probability 10 .1 0 – 10- .1 20 .15 10 – 20- .15 35 .4 20 – 35- .4 40 .3 35 – 40- .3 60 .05 40 – 60- .05 To use linear interpolation for continuous sampling, the discrete points on the end of each step need to be connected by line segments. This is represented in the graph below by the green line segments. The steps are represented in blue: rsample 60 50 40 30 20 10 0 x 0 .5 1 In the discrete case, sampling on step is accomplished by accumulating probabilities from the original table; e.g., for x = 0.4, accumulate probabilities until the cumulative probability exceeds 0.4; rsample is the event value at the point this happens (i.e., the cumulative probability 0.1+0.15+0.4 is the first to exceed 0.4, so the rsample value is 35). In the continuous case, the end points of the probability accumulation are needed, in this case x=0.25 and x=0.65 which represent the points (.25,20) and (.65,35) on the graph. -

Some Basic Probabilistic Processes
CHAPTER FOUR some basic probabilistic processes This chapter presents a few simple probabilistic processes and develops family relationships among the PMF's and PDF's associated with these processes. Although we shall encounter many of the most common PMF's and PDF's here, it is not our purpose to develop a general catalogue. A listing of the most frequently occurring PRfF's and PDF's and some of their properties appears as an appendix at the end of this book. 4-1 The 8er~ouil1Process A single Bernoulli trial generates an experimental value of discrete random variable x, described by the PMF to expand pkT(z) in a power series and then note the coefficient of zko in this expansion, recalling that any z transform may be written in the form pkT(~)= pk(0) + zpk(1) + z2pk(2) + ' ' This leads to the result known as the binomial PMF, 1-P xo-0 OlPll xo= 1 otherwise where the notation is the common Random variable x, as described above, is known as a Bernoulli random variable, and we note that its PMF has the z transform discussed in Sec. 1-9. Another way to derive the binomial PMF would be to work in a sequential sample space for an experiment which consists of n independ- The sample space for each Bernoulli trial is of the form ent Bernoulli trials, We have used the notation t success (;'') (;'') = Ifailure1 on the nth trial Either by use of the transform or by direct calculation we find We refer to the outcome of a Bernoulli trial as a success when the ex- Each sample point which represents an outcome of exactly ko suc- perimental value of x is unity and as a failure when the experimental cesses in the n trials would have a probability assignment equal to value of x is zero. -

Optimal Stopping Times for Estimating Bernoulli Parameters with Applications to Active Imaging
OPTIMAL STOPPING TIMES FOR ESTIMATING BERNOULLI PARAMETERS WITH APPLICATIONS TO ACTIVE IMAGING Safa C. Medin, John Murray-Bruce, and Vivek K Goyal Boston University Electrical and Computer Engineering Department ABSTRACT First we establish a framework for optimal adaptation of the number of trials for a single Bernoulli process. Then we consider a rectangu- We address the problem of estimating the parameter of a Bernoulli lar array of Bernoulli processes representing a scene in an imaging process. This arises in many applications, including photon-efficient problem, and we evaluate the inclusion of total variation regulariza- active imaging where each illumination period is regarded as a sin- tion for exploiting correlations among neighbors. In a simulation gle Bernoulli trial. We introduce a framework within which to min- with parameters realistic for active optical imaging, we demonstrate imize the mean-squared error (MSE) subject to an upper bound on a reduction in MSE by a factor of 2.67 (4.26 dB) in comparison to the mean number of trials. This optimization has several simple and the same regularized reconstruction approach applied without adap- intuitive properties when the Bernoulli parameter has a beta prior. In tation in numbers of trials. addition, by exploiting typical spatial correlation using total varia- tion regularization, we extend the developed framework to a rectan- gular array of Bernoulli processes representing the pixels in a natural 1.1. Related Work scene. In simulations inspired by realistic active imaging scenarios, we demonstrate a 4.26 dB reduction in MSE due to the adaptive In statistics, forming a parameter estimate from a number of i.i.d. -

Bernoulli Trials an Experiment, Or Trial, Whose Outcome Can Be Classified
Bernoulli trials An experiment, or trial, whose outcome can be classified as either a success or failure is performed. X = 1 when the outcome is a success 0 when outcome is a failure If p is the probability of a success then the pmf is, p(0) =P(X=0) =1-p p(1) =P(X=1) =p A random variable is called a Bernoulli random variable if it has the above pmf for p between 0 and 1. Expected value of Bernoulli r. v.: E(X) = 0*(1-p) + 1*p = p Variance of Bernoulli r. v.: E(X 2) = 02*(1-p) + 12*p = p Var(X) = E(X 2) - (E(X)) 2 = p - p2 = p(1-p) Ex. Flip a fair coin. Let X = number of heads. Then X is a Bernoulli random variable with p=1/2. E(X) = 1/2 Var(X) = 1/4 Binomial random variables Consider that n independent Bernoulli trials are performed. Each of these trials has probability p of success and probability (1-p) of failure. Let X = number of successes in the n trials. p(0) = P(0 successes in n trials) = (1-p)n {FFFFFFF} p(1) = P(1 success in n trials) = (n 1)p(1-p)n-1 {FSFFFFF} p(2) = P(2 successes in n trials) = (n 2)p2(1-p)n-2 {FSFSFFF} …… p(k) = P(k successes in n trials) = (n k)pk(1-p)n-k A random variable is called a Binomial(n,p) random variable if it has the pmf, p(k) = P(k successes in n trials) = (n k)pk(1-p)n-k for k=0,1,2,….n. -

Statistical Challenges with Modeling Motor Vehicle Crashes: Understanding the Implications of Alternative Approaches
STATISTICAL CHALLENGES WITH MODELING MOTOR VEHICLE CRASHES: UNDERSTANDING THE IMPLICATIONS OF ALTERNATIVE APPROACHES By Dominique Lord Associate Research Scientist Center for Transportation Safety Texas Transportation Institute Texas A&M University System 3135 TAMU, College Station, TX, 77843-3135 Tel. (979) 458-1218 E-mail: [email protected] Simon P. Washington Associate Professor Department of Civil Engineering & Engineering Mechanics University of Arizona Tucson, AZ 85721-0072 Tel. (520) 621- 4686 Email: [email protected] John N. Ivan Associate Professor Department of Civil & Environmental Engineering University of Connecticut 261 Glenbrook Road, Unit 2037 Storrs, CT 06269-2037 Tel. (860) 486-0352 E-mail: [email protected] March 22nd, 2004 Research Report Center for Transportation Safety Texas Transportation Institute Texas A&M University System 3135 TAMU, College Station, TX, 77843-3135 An earlier version of this paper was presented at the 83rd Annual Meeting of Transportation Research Board A shorter version of this paper with the new title “Poisson, Poisson-gamma and zero-inflated regression models of motor vehicle crashes: balancing statistical fit and theory” will be published in Accident Analysis & Prevention ABSTRACT There has been considerable research conducted over the last 20 years focused on predicting motor vehicle crashes on transportation facilities. The range of statistical models commonly applied includes binomial, Poisson, Poisson-gamma (or Negative Binomial), Zero-Inflated Poisson and Negative Binomial Models (ZIP and ZINB), and Multinomial probability models. Given the range of possible modeling approaches and the host of assumptions with each modeling approach, making an intelligent choice for modeling motor vehicle crash data is difficult at best. -

AP Statistics Chapter 8
Probability Models Chapter 16 Objectives: • Binomial Distribution – Conditions – Calculate binomial probabilities – Cumulative distribution function – Calculate means and standard deviations – Use normal approximation to the binomial distribution • Geometric Distribution – Conditions – Cumulative distribution function – Calculate means and standard deviations Bernoulli Trials • The basis for the probability models we will examine in this chapter is the Bernoulli trial. • We have Bernoulli trials if: – there are two possible outcomes (success and failure). – the probability of success, p, is constant. – the trials are independent. • Examples of Bernoulli Trials – Flipping a coin – Looking for defective products – Shooting free throws The Binomial Distribution The Binomial Model • A Binomial model tells us the probability for a random variable that counts the number of successes in a fixed number of Bernoulli trials. • Two parameters define the Binomial model: n, the number of trials; and, p, the probability of success. We denote this Binom(n, p). Independence • One of the important requirements for Bernoulli trials is that the trials be independent. • When we don’t have an infinite population, the trials are not independent. But, there is a rule that allows us to pretend we have independent trials: – The 10% condition: Bernoulli trials must be independent. If that assumption is violated, it is still okay to proceed as long as the sample is smaller than 10% of the population. Binomial Setting • Conditions required for a binomial distribution 1. Each observation falls into one of just two categories, “success” or “failure” (only two possible outcomes). 2. There are a fixed number n of observations. 3. The n observations are all independent. -

Chapter 4 Commonly Used Distributions Bernoulli Trial
Chapter 4 Commonly Used Distributions 4.1 Binomial distribution x3.4 Bernoulli trial: the simplest kind of random trial (like coin tossing) ♠ Each trial has two outcomes: S or F. ♠ For each trial, P (S) = p . ♠ The trials are independent. 76 ORF 245: Common Distributions { J.Fan 77 th Associated r.v.: let Yi be the outcome of i trial. Its takes only two values, f0; 1g, with pmf P (Yi = 1) = p and P (Yi = 0) = 1 − p, called Bernoulli dist. Then, E(Yi) = p and var(Yi) = pq. Generic name: |Quality control: Defective/Good; ♠Clinical trial: Survival/death; disease/non- disease; cure/not cure; effective/ineffective; |sampling survey: male/female; old/young; Coke/Pepsi; ♠online ads: click/not click .... Example 4.1 Which of following are Bernoulli trials? ♠ Roll a die 100 times and observe whether getting even or odd spots. ♠ Observe daily temperature and see whether it is above or below ORF 245: Common Distributions { J.Fan 78 freezing temperature. ♠ Sample 1000 people and observe whether they belong to upper, middle or low SES class. Definition: Let X be the number of successes in a Bernoulli process with n trials and probability of success p. Then, the dist of X called a Binomial distribution, denoted by X ∼ Bin(n; p). Fact: Let X ∼ Bin(n; p), then n x n−x • (pmf) b(x; n; p) = P (X = x) = x p q . • (summary) EX = np and var(X) = npq. • (sum of indep. r.v.) X = Y1 + ··· + Yn, FYi = a Bernoulli r.v. ORF 245: Common Distributions { J.Fan 79 Binomial with n= 8 p= 0.5 Binomial with n= 25 p= 0.5 Binomial with n= 50 p= 0.5 0.15 0.25 0.20 0.08 0.10 0.15