(Rubiaceae) and Related Species Using ISSR and RAPD Markers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Justus-Liebig-Universität Gießen Fachbereich Agrarwissenschaften
Justus‐Liebig‐Universität Gießen Fachbereich Agrarwissenschaften, Ökotrophologie und Umweltmanagement Institut für Landschaftsökologie und Ressourcenmanagement Professur für Landschaftsökologie und Landschaftsplanung Effects of Ground Cover on Seedling Emergence and Establishment Habilitationsschrift zur Erlangung der Lehrbefähigung für die Fächer „Vegetationsökologie und Landschaftsökologie“ im Fachbereich Agrarwissenschaften, Ökotrophologie und Umweltmanagement der Justus‐Liebig‐Universität Gießen vorgelegt von Dr. agr. Tobias W. Donath Gießen 2011 Justus Liebig University Gießen Faculty of Agricultural Sciences, Nutritional Sciences and Environmental Management Institute of Landscape Ecology and Resources Management Division of Landscape Ecology and Landscape Planning Effects of Ground Cover on Seedling Emergence and Establishment Habilitation Thesis for the Acquirement of the Facultas Docendi for the Subjects “Vegetation Ecology and Landscape Ecology” at the Faculty of Agricultural Sciences, Nutritional Sciences and Environmental Management Justus‐Liebig‐Universität Gießen Dr. agr. Tobias W. Donath Gießen 2011 Contents 1 Introduction ......................................................................................................................... 7 2 Interactions between litter and water availability affect seedling emergence in four familial pairs of floodplain species .................................................................... 15 Eckstein R. L. & Donath T. W. 2005 Journal of Ecology 93: 807‐816 3 Chemical effects -

EPPO Reporting Service
ORGANISATION EUROPEENNE EUROPEAN AND MEDITERRANEAN ET MEDITERRANEENNE PLANT PROTECTION POUR LA PROTECTION DES PLANTES ORGANIZATION EPPO Reporting Service NO. 04 PARIS, 2013-04-01 CONTENTS 2013/075 - Appointment of a new Director-General of EPPO _______________________________________________________________________ Pests & Diseases 2013/076 - Anoplophora glabripennis eradicated from Canada 2013/077 - Anoplophora glabripennis eradicated from New Jersey (US) 2013/078 - First report of Tuta absoluta in the United Arab Emirates 2013/079 - First report of Tuta absoluta in Yemen 2013/080 - First report of Globodera pallida in Denmark 2013/081 - First report of Helicoverpa armigera in Brazil 2013/082 - Eradication of Clavibacter michiganensis subsp. sepedonicus from Denmark 2013/083 - First report of Pantoea stewartii in Argentina 2013/084 - First report of ‘Candidatus Phytoplasma mali’ in Canada 2013/085 - 'Candidatus Phytoplasma solani' formally proposed as a new phytoplasma species 2013/086 - First report of Tomato spotted wilt virus in Bosnia and Herzegovina 2013/087 - New data on quarantine pests and pests of the EPPO Alert List 2013/088 - EPPO report on notifications of non-compliance CONTENTS ________________________________________________________________________ Invasive Plants 2013/089 - Invasive alien plants in Finland 2013/090 - Update on the world distribution of Asparagus asparagoides 2013/091 - Eradication of Asparagus asparagoides in Andalucia (ES) 2013/092 - Cardiospermum grandiflorum in Macaronesia 2013/093 - Revision of ISPM -

Medicinal and Aromatic Plants of Azerbaijan – Naiba Mehtiyeva and Sevil Zeynalova
ETHNOPHARMACOLOGY – Medicinal and Aromatic Plants of Azerbaijan – Naiba Mehtiyeva and Sevil Zeynalova MEDICINAL AND AROMATIC PLANTS OF AZERBAIJAN Naiba Mehtiyeva and Sevil Zeynalova Institute of Botany, Azerbaijan National Academy of Sciences, Badamdar sh. 40, AZ1073, Baku, Azerbaijan Keywords: Azerbaijan, medicinal plants, aromatic plants, treatments, history, biological active substances. Contents 1. Introduction 2. Historical perspective of the traditional medicine 3. Medicinal and aromatic plants of Azerbaijan 4. Preparation and applying of decoctions and infusions from medicinal plants 5. Conclusion Acknowledgement Bibliography Biographical Sketches Summary Data on the biological active substances and therapeutical properties of more than 131 medicinal and aromatic (spicy-aromatic) plants widely distributed and frequently used in Azerbaijan are given in this chapter. The majority of the described species contain flavonoids (115 sp.), vitamin C (84 sp.), fatty oils (78 sp.), tannins (77 sp.), alkaloids (74 sp.) and essential oils (73 sp.). A prevalence of these biological active substances defines the broad spectrum of therapeutic actions of the described plants. So, significant number of species possess antibacterial (69 sp.), diuretic (60 sp.), wound healing (51 sp.), styptic (46 sp.) and expectorant (45 sp.) peculiarities. The majority of the species are used in curing of gastrointestinal (89 sp.), bronchopulmonary (61 sp.), dermatovenerologic (61 sp.), nephritic (55 sp.) and infectious (52 sp.) diseases, also for treatment of festering -

Effective Typification of Galium Verum Var. Hallaensis, a Replacement Name for the Korean G
Phytotaxa 423 (5): 289–292 ISSN 1179-3155 (print edition) https://www.mapress.com/j/pt/ PHYTOTAXA Copyright © 2019 Magnolia Press Correspondence ISSN 1179-3163 (online edition) https://doi.org/10.11646/phytotaxa.423.5.3 Effective typification of Galium verum var. hallaensis, a replacement name for the Korean G. pusillum (Rubiaceae) DONG CHAN SON1 & KAE SUN CHANG1* 1Division of Forest Biodiversity and Herbarium, Korea National Arboretum, Pocheon-si, Gyeonggi-do 11186, Republic of Korea *Author for correspondence: [email protected] The genus Galium Linnaeus (1753: 105), a well-known member of the madder family, consists of around 650 perennial and annual herbaceous species widely distributed in temperate, subtropical, and tropical regions worldwide (Ehrendorfer et al. 2005, Chen & Ehrendorfer 2011). This genus is distinguished from other genera belonging to Rubiaceae by several morphological characteristics including the leaves in whorls of 2–8, 3 or 4-lobed corollas, rudimentary corolla tubes, and dry mericarps (Yamazaki 1993, Ehrendorfer et al. 2014, Elkordy & Schanzer 2015). Article 9.23 of the Shenzhen Code (Turland et al. 2018) requires that “on or after 1 January 2001, lectotypification, neotypification, or epitypification of a name of a species or infraspecific taxon is not effected unless indicated by use of the term “lectotypus”, “neotypus”, or “epitypus”, its abbreviation, or its equivalent in a modern language,” and Art. 7.11 requires that “… designation of a type is achieved only…, on or after 1 January 2001, if the typification statement includes the phrase “designated here” (hic designatus) or an equivalent.” However, in many monographic and taxonomic works published on or after 1 January 2001, the requirements of Art. -

28. GALIUM Linnaeus, Sp. Pl. 1: 105. 1753
Fl. China 19: 104–141. 2011. 28. GALIUM Linnaeus, Sp. Pl. 1: 105. 1753. 拉拉藤属 la la teng shu Chen Tao (陈涛); Friedrich Ehrendorfer Subshrubs to perennial or annual herbs. Stems often weak and clambering, often notably prickly or “sticky” (i.e., retrorsely aculeolate, “velcro-like”). Raphides present. Leaves opposite, mostly with leaflike stipules in whorls of 4, 6, or more, usually sessile or occasionally petiolate, without domatia, abaxial epidermis sometimes punctate- to striate-glandular, mostly with 1 main nerve, occasionally triplinerved or palmately veined; stipules interpetiolar and usually leaflike, sometimes reduced. Inflorescences mostly terminal and axillary (sometimes only axillary), thyrsoid to paniculiform or subcapitate, cymes several to many flowered or in- frequently reduced to 1 flower, pedunculate to sessile, bracteate or bracts reduced especially on higher order axes [or bracts some- times leaflike and involucral], bracteoles at pedicels lacking. Flowers mostly bisexual and monomorphic, hermaphroditic, sometimes unisexual, andromonoecious, occasionally polygamo-dioecious or dioecious, pedicellate to sessile, usually quite small. Calyx with limb nearly always reduced to absent; hypanthium portion fused with ovary. Corolla white, yellow, yellow-green, green, more rarely pink, red, dark red, or purple, rotate to occasionally campanulate or broadly funnelform; tube sometimes so reduced as to give appearance of free petals, glabrous inside; lobes (3 or)4(or occasionally 5), valvate in bud. Stamens (3 or)4(or occasionally 5), inserted on corolla tube near base, exserted; filaments developed to ± reduced; anthers dorsifixed. Inferior ovary 2-celled, ± didymous, ovoid, ellipsoid, or globose, smooth, papillose, tuberculate, or with hooked or rarely straight trichomes, 1 erect and axile ovule in each cell; stigmas 2-lobed, exserted. -

Cambridgeshire and Peterborough County Wildlife Sites
Cambridgeshire and Peterborough County Wildlife Sites Selection Guidelines VERSION 6.2 April 2014 CAMBRIDGESHIRE & PETERBOROUGH COUNTY WILDLIFE SITES PANEL CAMBRIDGESHIRE & PETERBOROUGH COUNTY WILDLIFE SITES PANEL operates under the umbrella of the Cambridgeshire and Peterborough Biodiversity Partnership. The panel includes suitably qualified and experienced representatives from The Wildlife Trust for Bedfordshire, Cambridgeshire, Northamptonshire; Natural England; The Environment Agency; Cambridgeshire County Council; Peterborough City Council; South Cambridgeshire District Council; Huntingdonshire District Council; East Cambridgeshire District Council; Fenland District Council; Cambridgeshire and Peterborough Environmental Records Centre and many amateur recorders and recording groups. Its aim is to agree the basis for site selection, reviewing and amending them as necessary based on the best available biological information concerning the county. © THE WILDLIFE TRUST FOR BEDFORDSHIRE, CAMBRIDGESHIRE AND NORTHAMPTONSHIRE 2014 © Appendices remain the copyright of their respective originators. All rights reserved. Without limiting the rights under copyright reserved above, no part of this publication may be reproduced, stored in any type of retrieval system or transmitted in any form or by any means (electronic, photocopying, mechanical, recording or otherwise) without the permission of the copyright owner. INTRODUCTION The Selection Criteria are substantially based on Guidelines for selection of biological SSSIs published by the Nature Conservancy Council (succeeded by English Nature) in 1989. Appropriate modifications have been made to accommodate the aim of selecting a lower tier of sites, i.e. those sites of county and regional rather than national importance. The initial draft has been altered to reflect the views of the numerous authorities consulted during the preparation of the Criteria and to incorporate the increased knowledge of the County's habitat resource gained by the Phase 1 Habitat Survey (1992-97) and other survey work in the past decade. -

Determining the Emergence Timing, Morphological Characteristics, and Species Composition of Galium Populations in Western Canada
Determining the emergence timing, morphological characteristics, and species composition of Galium populations in western Canada A Thesis Submitted to the College of Graduate Studies and Research In Partial fulfillment of the Requirements For the Degree of Master of Science In the Department of Plant Sciences University of Saskatchewan Saskatoon By Andrea C. De Roo © Copyright Andrea C. De Roo, 2016. All rights reserved. Permission to Use In presenting this thesis in partial fulfillment of the requirements for a Postgraduate degree from the University of Saskatchewan, I agree that the Libraries of this University may make it freely available for inspection. I further agree that permission for copying of this thesis in any manner, in whole or in part, for scholarly purpose may be granted by the professor or professors who supervised my thesis work or, in their absence, by the Head of the Department or the Dean of the College in which my thesis work was done. It is understood that any copying, publication, or use of this thesis or parts thereof for financial gain shall not be allowed without my written permission. It is also understood that due recognition shall be given to me and to the University of Saskatchewan in any scholarly use which may be made of any material in my thesis. Request for permission to copy or to make other use of material in this thesis, in whole or part, should be addressed to: Head of the Department of Plant Sciences 51 Campus Drive University of Saskatchewan Saskatoon, Saskatchewan (S7N 5A8) i Abstract Three species of Galium are commonly believed to thrive in western Canada; Galium aparine L., Galium spurium L. -

Aegopodium Podagraria
Aegopodium podagraria INTRODUCTORY DISTRIBUTION AND OCCURRENCE BOTANICAL AND ECOLOGICAL CHARACTERISTICS FIRE EFFECTS AND MANAGEMENT MANAGEMENT CONSIDERATIONS APPENDIX: FIRE REGIME TABLE REFERENCES INTRODUCTORY AUTHORSHIP AND CITATION FEIS ABBREVIATION NRCS PLANT CODE COMMON NAMES TAXONOMY SYNONYMS LIFE FORM Variegated goutweed. All-green goutweed. Photos by John Randall, The Nature Conservancy, Bugwood.org AUTHORSHIP AND CITATION: Waggy, Melissa, A. 2010. Aegopodium podagraria. In: Fire Effects Information System, [Online]. U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station, Fire Sciences Laboratory (Producer). Available: http://www.fs.fed.us/database/feis/ [ 2010, January 21]. FEIS ABBREVIATION: AEGPOD NRCS PLANT CODE [87]: AEPO COMMON NAMES: goutweed bishop's goutweed bishop's weed bishopsweed ground elder herb Gerard TAXONOMY: The scientific name of goutweed is Aegopodium podagraria L. (Apiaceae) [40]. SYNONYMS: Aegopodium podagraria var. podagraria [71] Aegopodium podagraria var. variegatum Bailey [40,71] LIFE FORM: Forb DISTRIBUTION AND OCCURRENCE SPECIES: Aegopodium podagraria GENERAL DISTRIBUTION HABITAT TYPES AND PLANT COMMUNITIES GENERAL DISTRIBUTION: Goutweed was introduced in North America from Europe [82]. In the United States, goutweed occurs from Maine south to South Carolina and west to Minnesota and Missouri. It also occurs in the Pacific Northwest from Montana to Washington and Oregon. It occurs in all the Canadian provinces excepting Newfoundland and Labrador, and Alberta. Plants Database provides a distributional map of goutweed. Globally, goutweed occurs primarily in the northern hemisphere, particularly in Europe, Asia Minor ([28,36,58,92], reviews by [14,27]), and Russia (review by [27,63]). Goutweed's native distribution is unclear. It may have been introduced in England (review by [2]) and is considered a "weed" in the former Soviet Union, Germany, Finland (Holm 1979 cited in [14]), and Poland [44]. -
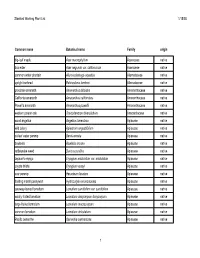
Plant List for Web Page
Stanford Working Plant List 1/15/08 Common name Botanical name Family origin big-leaf maple Acer macrophyllum Aceraceae native box elder Acer negundo var. californicum Aceraceae native common water plantain Alisma plantago-aquatica Alismataceae native upright burhead Echinodorus berteroi Alismataceae native prostrate amaranth Amaranthus blitoides Amaranthaceae native California amaranth Amaranthus californicus Amaranthaceae native Powell's amaranth Amaranthus powellii Amaranthaceae native western poison oak Toxicodendron diversilobum Anacardiaceae native wood angelica Angelica tomentosa Apiaceae native wild celery Apiastrum angustifolium Apiaceae native cutleaf water parsnip Berula erecta Apiaceae native bowlesia Bowlesia incana Apiaceae native rattlesnake weed Daucus pusillus Apiaceae native Jepson's eryngo Eryngium aristulatum var. aristulatum Apiaceae native coyote thistle Eryngium vaseyi Apiaceae native cow parsnip Heracleum lanatum Apiaceae native floating marsh pennywort Hydrocotyle ranunculoides Apiaceae native caraway-leaved lomatium Lomatium caruifolium var. caruifolium Apiaceae native woolly-fruited lomatium Lomatium dasycarpum dasycarpum Apiaceae native large-fruited lomatium Lomatium macrocarpum Apiaceae native common lomatium Lomatium utriculatum Apiaceae native Pacific oenanthe Oenanthe sarmentosa Apiaceae native 1 Stanford Working Plant List 1/15/08 wood sweet cicely Osmorhiza berteroi Apiaceae native mountain sweet cicely Osmorhiza chilensis Apiaceae native Gairdner's yampah (List 4) Perideridia gairdneri gairdneri Apiaceae -

Bonpland and Humboldt Specimens, Field Notes, and Herbaria; New Insights from a Study of the Monocotyledons Collected in Venezuela
Bonpland and Humboldt specimens, field notes, and herbaria; new insights from a study of the monocotyledons collected in Venezuela Fred W. Stauffer, Johann Stauffer & Laurence J. Dorr Abstract Résumé STAUFFER, F. W., J. STAUFFER & L. J. DORR (2012). Bonpland and STAUFFER, F. W., J. STAUFFER & L. J. DORR (2012). Echantillons de Humboldt specimens, field notes, and herbaria; new insights from a study Bonpland et Humboldt, carnets de terrain et herbiers; nouvelles perspectives of the monocotyledons collected in Venezuela. Candollea 67: 75-130. tirées d’une étude des monocotylédones récoltées au Venezuela. Candollea In English, English and French abstracts. 67: 75-130. En anglais, résumés anglais et français. The monocotyledon collections emanating from Humboldt and Les collections de Monocotylédones provenant des expéditions Bonpland’s expedition are used to trace the complicated ways de Humboldt et Bonpland sont utilisées ici pour retracer les in which botanical specimens collected by the expedition were cheminements complexes des spécimens collectés lors returned to Europe, to describe the present location and to de leur retour en Europe. Ces collections sont utilisées pour explore the relationship between specimens, field notes, and établir la localisation actuelle et la composition d’importants descriptions published in the multi-volume “Nova Genera et jeux de matériel associés à ce voyage, ainsi que pour explorer Species Plantarum” (1816-1825). Collections in five European les relations existantes entre les spécimens, les notes de terrain herbaria were searched for monocotyledons collected by et les descriptions parues dans les divers volumes de «Nova the explorers. In Paris, a search of the Bonpland Herbarium Genera et Species Plantarum» (1816-1825). -

Flora Mediterranea 26
FLORA MEDITERRANEA 26 Published under the auspices of OPTIMA by the Herbarium Mediterraneum Panormitanum Palermo – 2016 FLORA MEDITERRANEA Edited on behalf of the International Foundation pro Herbario Mediterraneo by Francesco M. Raimondo, Werner Greuter & Gianniantonio Domina Editorial board G. Domina (Palermo), F. Garbari (Pisa), W. Greuter (Berlin), S. L. Jury (Reading), G. Kamari (Patras), P. Mazzola (Palermo), S. Pignatti (Roma), F. M. Raimondo (Palermo), C. Salmeri (Palermo), B. Valdés (Sevilla), G. Venturella (Palermo). Advisory Committee P. V. Arrigoni (Firenze) P. Küpfer (Neuchatel) H. M. Burdet (Genève) J. Mathez (Montpellier) A. Carapezza (Palermo) G. Moggi (Firenze) C. D. K. Cook (Zurich) E. Nardi (Firenze) R. Courtecuisse (Lille) P. L. Nimis (Trieste) V. Demoulin (Liège) D. Phitos (Patras) F. Ehrendorfer (Wien) L. Poldini (Trieste) M. Erben (Munchen) R. M. Ros Espín (Murcia) G. Giaccone (Catania) A. Strid (Copenhagen) V. H. Heywood (Reading) B. Zimmer (Berlin) Editorial Office Editorial assistance: A. M. Mannino Editorial secretariat: V. Spadaro & P. Campisi Layout & Tecnical editing: E. Di Gristina & F. La Sorte Design: V. Magro & L. C. Raimondo Redazione di "Flora Mediterranea" Herbarium Mediterraneum Panormitanum, Università di Palermo Via Lincoln, 2 I-90133 Palermo, Italy [email protected] Printed by Luxograph s.r.l., Piazza Bartolomeo da Messina, 2/E - Palermo Registration at Tribunale di Palermo, no. 27 of 12 July 1991 ISSN: 1120-4052 printed, 2240-4538 online DOI: 10.7320/FlMedit26.001 Copyright © by International Foundation pro Herbario Mediterraneo, Palermo Contents V. Hugonnot & L. Chavoutier: A modern record of one of the rarest European mosses, Ptychomitrium incurvum (Ptychomitriaceae), in Eastern Pyrenees, France . 5 P. Chène, M. -

Connaissance De La Flore Rare Ou Menacée De Franche-Comté C Asperula Tinctoria L
Connaissance de la flore rare ou menacée de Franche-Comté C Asperula tinctoria L. B F C ASSOCI A TION LOI 1901 PORTE RIVOTTE 25000 BESANÇON TE L /F A X : 03 81 83 03 58 E-MA I L : [email protected] Décembre 2006 Conservatoire Botanique de Franche-Comté FERREZ Y., 2004. Connaissance de la flore rare ou menacée de Franche-Comté, Asperula tinctoria L., Conservatoire Botanique de Franche-Comté, 26 p. Cliché de couverture : Asperula tinctoria L., FERREZ Y., 2006 2 Connaissance de la flore rare ou menacée de Franche-Comté, Asperula tinctoria L. CONSERVATOIRE BOTANIQUE DE FRAN C HE -COMTÉ Connaissance de la flore rare ou menacée de Franche-Comté Asperula tinctoria L. Décembre 2006 Etude réalisée par le Conservatoire Inventaires de terrain : YORICK FERREZ , Botanique de Franche-Comté. CHRISTOPHE HENNEQUIN pour le compte de la Direction Régionale de Analyse des données : YORICK FERREZ l’Environnement de Franche-Comté et du Conseil Régional de Franche-Comté. Rédaction et mise en page : YORICK FERREZ Relecture : FRANÇOIS DEHON D T , PASCALE NUSSBAUM Connaissance de la flore rare ou menacée de Franche-Comté, Asperula tinctoria L. Sommaire 1 - Données générales sur l’espèce 3 1.1 - Nomenclature 3 1.2 - Traits distinctifs 3 1.3 - Biologie et particularité du taxon 4 1.4 - Répartition générale et menaces 4 1.5 - Statut de protection et de menace 4 2 - Statut du taxon et situation actuelle en Franche-Comté 4 2.1 - Données historiques (antérieures à 1964) 4 2.2 - Données anciennes (antérieures à 1984) 5 2.3 - Données récentes (postérieures à 1985) 5