Assessing Sampling Coverage of Species Distribution in Biodiversity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Flowering Plants Eudicots Apiales, Gentianales (Except Rubiaceae)
Edited by K. Kubitzki Volume XV Flowering Plants Eudicots Apiales, Gentianales (except Rubiaceae) Joachim W. Kadereit · Volker Bittrich (Eds.) THE FAMILIES AND GENERA OF VASCULAR PLANTS Edited by K. Kubitzki For further volumes see list at the end of the book and: http://www.springer.com/series/1306 The Families and Genera of Vascular Plants Edited by K. Kubitzki Flowering Plants Á Eudicots XV Apiales, Gentianales (except Rubiaceae) Volume Editors: Joachim W. Kadereit • Volker Bittrich With 85 Figures Editors Joachim W. Kadereit Volker Bittrich Johannes Gutenberg Campinas Universita¨t Mainz Brazil Mainz Germany Series Editor Prof. Dr. Klaus Kubitzki Universita¨t Hamburg Biozentrum Klein-Flottbek und Botanischer Garten 22609 Hamburg Germany The Families and Genera of Vascular Plants ISBN 978-3-319-93604-8 ISBN 978-3-319-93605-5 (eBook) https://doi.org/10.1007/978-3-319-93605-5 Library of Congress Control Number: 2018961008 # Springer International Publishing AG, part of Springer Nature 2018 This work is subject to copyright. All rights are reserved by the Publisher, whether the whole or part of the material is concerned, specifically the rights of translation, reprinting, reuse of illustrations, recitation, broadcasting, reproduction on microfilms or in any other physical way, and transmission or information storage and retrieval, electronic adaptation, computer software, or by similar or dissimilar methodology now known or hereafter developed. The use of general descriptive names, registered names, trademarks, service marks, etc. in this publication does not imply, even in the absence of a specific statement, that such names are exempt from the relevant protective laws and regulations and therefore free for general use. -

Assessing Sampling Coverage of Species Distribution in Biodiversity Databases
1 Assessing sampling coverage of species distribution in biodiversity 2 databases 3 4 Running title: Sampling coverage by box-counting 5 6 Maria Sporbert 1,2 *, Helge Bruelheide 1,2, Gunnar Seidler 1, Petr Keil 1,3, Ute Jandt 1,2, Gunnar 7 Austrheim 4, Idoia Biurrun 5, Juan Antonio Campos 5, Andraž Čarni 6,7, Milan Chytrý 8, János Csiky 9, 8 Els De Bie 10, Jürgen Dengler 2,11,12, Valentin Golub 13, John-Arvid Grytnes 14, Adrian Indreica 15, 9 Florian Jansen 16, Martin Jiroušek 8,17, Jonathan Lenoir 18, Miska Luoto 19, Corrado Marcenò 5, Jesper 10 Erenskjold Moeslund 20, Aaron Pérez-Haase 21, Solvita Rūsiņa 22, Vigdis Vandvik 23,24, Kiril Vassilev 11 25, Erik Welk 1,2 12 13 1Institute of Biology / Geobotany and Botanical Garden, Martin Luther University Halle-Wittenberg, 14 Halle, Germany 15 2German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, Leipzig, Germany 16 3Institute of Computer Science / Biodiversity Synthesis, Martin Luther University Halle-Wittenberg, 17 Halle, Germany 18 4Department of Natural History, University Museum Norwegian University of Science and 19 Technology, Trondheim, Norway 20 5Department Plant Biology and Ecology, University of the Basque Country UPV/EHU, Bilbao, Spain 21 6Scientific Research Centre of the Slovenian Academy of Sciences and Arts, Jovan Hadži Institute of 22 Biology, Ljubljana, Slovenia 23 7School for Viticulture and Enology, University of Nova Gorica, Nova Gorica, Slovenia 24 8Department of Botany and Zoology, Faculty of Science, Masaryk University, Brno, Czech Republic -

Monitoring Methodology and Protocols for 20 Habitats, 20 Species and 20 Birds
1 Finnish Environment Institute SYKE, Finland Monitoring methodology and protocols for 20 habitats, 20 species and 20 birds Twinning Project MK 13 IPA EN 02 17 Strengthening the capacities for effective implementation of the acquis in the field of nature protection Report D 3.1. - 1. 7.11.2019 Funded by the European Union The Ministry of Environment and Physical Planning, Department of Nature, Republic of North Macedonia Metsähallitus (Parks and Wildlife Finland), Finland The State Service for Protected Areas (SSPA), Lithuania 2 This project is funded by the European Union This document has been produced with the financial support of the European Union. Its contents are the sole responsibility of the Twinning Project MK 13 IPA EN 02 17 and and do not necessarily reflect the views of the European Union 3 Table of Contents 1. Introduction .......................................................................................................................................................... 6 Summary 6 Overview 8 Establishment of Natura 2000 network and the process of site selection .............................................................. 9 Preparation of reference lists for the species and habitats ..................................................................................... 9 Needs for data .......................................................................................................................................................... 9 Protocols for the monitoring of birds .................................................................................................................... -

Conserving Europe's Threatened Plants
Conserving Europe’s threatened plants Progress towards Target 8 of the Global Strategy for Plant Conservation Conserving Europe’s threatened plants Progress towards Target 8 of the Global Strategy for Plant Conservation By Suzanne Sharrock and Meirion Jones May 2009 Recommended citation: Sharrock, S. and Jones, M., 2009. Conserving Europe’s threatened plants: Progress towards Target 8 of the Global Strategy for Plant Conservation Botanic Gardens Conservation International, Richmond, UK ISBN 978-1-905164-30-1 Published by Botanic Gardens Conservation International Descanso House, 199 Kew Road, Richmond, Surrey, TW9 3BW, UK Design: John Morgan, [email protected] Acknowledgements The work of establishing a consolidated list of threatened Photo credits European plants was first initiated by Hugh Synge who developed the original database on which this report is based. All images are credited to BGCI with the exceptions of: We are most grateful to Hugh for providing this database to page 5, Nikos Krigas; page 8. Christophe Libert; page 10, BGCI and advising on further development of the list. The Pawel Kos; page 12 (upper), Nikos Krigas; page 14: James exacting task of inputting data from national Red Lists was Hitchmough; page 16 (lower), Jože Bavcon; page 17 (upper), carried out by Chris Cockel and without his dedicated work, the Nkos Krigas; page 20 (upper), Anca Sarbu; page 21, Nikos list would not have been completed. Thank you for your efforts Krigas; page 22 (upper) Simon Williams; page 22 (lower), RBG Chris. We are grateful to all the members of the European Kew; page 23 (upper), Jo Packet; page 23 (lower), Sandrine Botanic Gardens Consortium and other colleagues from Europe Godefroid; page 24 (upper) Jože Bavcon; page 24 (lower), Frank who provided essential advice, guidance and supplementary Scumacher; page 25 (upper) Michael Burkart; page 25, (lower) information on the species included in the database. -
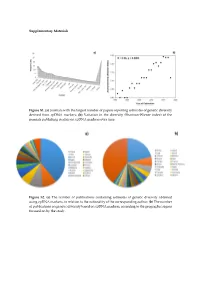
(A) Journals with the Largest Number of Papers Reporting Estimates Of
Supplementary Materials Figure S1. (a) Journals with the largest number of papers reporting estimates of genetic diversity derived from cpDNA markers; (b) Variation in the diversity (Shannon-Wiener index) of the journals publishing studies on cpDNA markers over time. Figure S2. (a) The number of publications containing estimates of genetic diversity obtained using cpDNA markers, in relation to the nationality of the corresponding author; (b) The number of publications on genetic diversity based on cpDNA markers, according to the geographic region focused on by the study. Figure S3. Classification of the angiosperm species investigated in the papers that analyzed genetic diversity using cpDNA markers: (a) Life mode; (b) Habitat specialization; (c) Geographic distribution; (d) Reproductive cycle; (e) Type of flower, and (f) Type of pollinator. Table S1. Plant species identified in the publications containing estimates of genetic diversity obtained from the use of cpDNA sequences as molecular markers. Group Family Species Algae Gigartinaceae Mazzaella laminarioides Angiospermae Typhaceae Typha laxmannii Angiospermae Typhaceae Typha orientalis Angiospermae Typhaceae Typha angustifolia Angiospermae Typhaceae Typha latifolia Angiospermae Araliaceae Eleutherococcus sessiliflowerus Angiospermae Polygonaceae Atraphaxis bracteata Angiospermae Plumbaginaceae Armeria pungens Angiospermae Aristolochiaceae Aristolochia kaempferi Angiospermae Polygonaceae Atraphaxis compacta Angiospermae Apocynaceae Lagochilus macrodontus Angiospermae Polygonaceae Atraphaxis -

A Review of European Progress Towards the Global Strategy for Plant Conservation 2011-2020
A review of European progress towards the Global Strategy for Plant Conservation 2011-2020 1 A review of European progress towards the Global Strategy for Plant Conservation 2011-2020 The geographical area of ‘Europe’ includes the forty seven countries of the Council of Europe and Belarus: Albania, Andorra, Armenia, Austria, Azerbaijan, Belarus, Belgium, Bosnia-Herzegovina, Bulgaria, Croatia, Cyprus, Czech Republic, Denmark, Estonia, Finland, France, Georgia, Germany, Greece, Hungary, Iceland, Ireland, Italy, Latvia, Liechtenstein, Lithuania, Luxembourg, Malta, Republic of Moldova, Monaco, Montenegro, Netherlands, North Macedonia, Norway, Poland, Portugal, Romania, Russian Federation, San Marino, Serbia, Slovakia, Slovenia, Spain, Sweden, Switzerland, Turkey, Ukraine, United Kingdom. Front Cover Image: Species rich meadow with Papaver paucifoliatum, Armenia, Anna Asatryan. Disclaimer: The designations employed and the presentation of material in this publication do not imply the expression of any opinion whatsoever on the part of the copyright holders concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. The mentioning of specific companies or products does not imply that they are endorsed or recommended by PLANTA EUROPA or Plantlife International or preferred to others that are not mentioned – they are simply included as examples. All reasonable precautions have been taken by PLANTA EUROPA and Plantlife International to verify the information contained in this publication. However, the published material is being distributed without warranty of any kind, either expressed or implied. The responsibility for the interpretation and use of the material lies with the reader. In no event shall PLANTA EUROPA, Plantlife International or the authors be liable for any consequences whatsoever arising from its use. -

Evolutionary Shifts in Fruit Dispersal Syndromes in Apiaceae Tribe Scandiceae
Plant Systematics and Evolution (2019) 305:401–414 https://doi.org/10.1007/s00606-019-01579-1 ORIGINAL ARTICLE Evolutionary shifts in fruit dispersal syndromes in Apiaceae tribe Scandiceae Aneta Wojewódzka1,2 · Jakub Baczyński1 · Łukasz Banasiak1 · Stephen R. Downie3 · Agnieszka Czarnocka‑Cieciura1 · Michał Gierek1 · Kamil Frankiewicz1 · Krzysztof Spalik1 Received: 17 November 2018 / Accepted: 2 April 2019 / Published online: 2 May 2019 © The Author(s) 2019 Abstract Apiaceae tribe Scandiceae includes species with diverse fruits that depending upon their morphology are dispersed by gravity, carried away by wind, or transported attached to animal fur or feathers. This diversity is particularly evident in Scandiceae subtribe Daucinae, a group encompassing species with wings or spines developing on fruit secondary ribs. In this paper, we explore fruit evolution in 86 representatives of Scandiceae and outgroups to assess adaptive shifts related to the evolutionary switch between anemochory and epizoochory and to identify possible dispersal syndromes, i.e., patterns of covariation of morphological and life-history traits that are associated with a particular vector. We also assess the phylogenetic signal in fruit traits. Principal component analysis of 16 quantitative fruit characters and of plant height did not clearly separate spe- cies having diferent dispersal strategies as estimated based on fruit appendages. Only presumed anemochory was weakly associated with plant height and the fattening of mericarps with their accompanying anatomical changes. We conclude that in Scandiceae, there are no distinct dispersal syndromes, but a continuum of fruit morphologies relying on diferent dispersal vectors. Phylogenetic mapping of ten discrete fruit characters on trees inferred by nrDNA ITS and cpDNA sequence data revealed that all are homoplastic and of limited use for the delimitation of genera. -

Desktop Biodiversity Report
Desktop Biodiversity Report Land at Balcombe Parish ESD/14/747 Prepared for Katherine Daniel (Balcombe Parish Council) 13th February 2014 This report is not to be passed on to third parties without prior permission of the Sussex Biodiversity Record Centre. Please be aware that printing maps from this report requires an appropriate OS licence. Sussex Biodiversity Record Centre report regarding land at Balcombe Parish 13/02/2014 Prepared for Katherine Daniel Balcombe Parish Council ESD/14/74 The following information is included in this report: Maps Sussex Protected Species Register Sussex Bat Inventory Sussex Bird Inventory UK BAP Species Inventory Sussex Rare Species Inventory Sussex Invasive Alien Species Full Species List Environmental Survey Directory SNCI M12 - Sedgy & Scott's Gills; M22 - Balcombe Lake & associated woodlands; M35 - Balcombe Marsh; M39 - Balcombe Estate Rocks; M40 - Ardingly Reservior & Loder Valley Nature Reserve; M42 - Rowhill & Station Pastures. SSSI Worth Forest. Other Designations/Ownership Area of Outstanding Natural Beauty; Environmental Stewardship Agreement; Local Nature Reserve; National Trust Property. Habitats Ancient tree; Ancient woodland; Ghyll woodland; Lowland calcareous grassland; Lowland fen; Lowland heathland; Traditional orchard. Important information regarding this report It must not be assumed that this report contains the definitive species information for the site concerned. The species data held by the Sussex Biodiversity Record Centre (SxBRC) is collated from the biological recording community in Sussex. However, there are many areas of Sussex where the records held are limited, either spatially or taxonomically. A desktop biodiversity report from SxBRC will give the user a clear indication of what biological recording has taken place within the area of their enquiry. -

Samenkatalog Graz 2016.Pdf
SAMENTAUSCHVERZEICHNIS Index Seminum Seed list Catalogue de graines des Botanischen Gartens der Karl-Franzens-Universität Graz Ernte / Harvest / Récolte 2016 Herausgegeben von Christian BERG, Kurt MARQUART & Jonathan WILFLING ebgconsortiumindexseminum2012 Institut für Pflanzenwissenschaften, Januar 2017 Botanical Garden, Institute of Plant Sciences, Karl- Franzens-Universität Graz 2 Botanischer Garten Institut für Pflanzenwissenschaften Karl-Franzens-Universität Graz Holteigasse 6 A - 8010 Graz, Austria Fax: ++43-316-380-9883 Email- und Telefonkontakt: [email protected], Tel.: ++43-316-380-5651 [email protected], Tel.: ++43-316-380-5747 Webseite: http://garten.uni-graz.at/ Zitiervorschlag : BERG, C., MARQUART, K. & Wilfling, J. (2017): Samentauschverzeichnis – Index Seminum – des Botanischen Gartens der Karl-Franzens-Universität Graz, Samenernte 2016. – 54 S., Karl-Franzens-Universität Graz. Personalstand des Botanischen Gartens Graz: Institutsleiter: Ao. Univ.-Prof. Mag. Dr. Helmut MAYRHOFER Wissenschaftlicher Gartenleiter: Dr. Christian BERG Gartenverwalter: Jonathan WILFLING, B. Sc. Gärtnermeister: Friedrich STEFFAN GärtnerInnen: Doris ADAM-LACKNER Viola BONGERS Magarete HIDEN Franz HÖDL Kurt MARQUART Franz STIEBER Ulrike STRAUSSBERGER Monika GABER Gartenarbeiter: Philip FRIEDL René MICHALSKI Oliver KROPIWNICKI Gärtnerlehrlinge: Gabriel Buchmann (1. Lehrjahr) Bahram EMAMI (3. Lehrjahr) Mario MARX (3. Lehrjahr) 3 Inhaltsverzeichnis / Contents / Table des matières Abkürzungen / List of abbreviations / Abréviations -

Central European Vegetation
Plant Formations in the Central European BioProvince Peter Martin Rhind Central European Beech Woodlands Beech (Fagus sylvatica) woods form the natural climax over much of Central Europe where the soils are relatively dry and can extend well into the uplands in the more southern zones. In the north, however, around Sweden it is confined to the lowlands. Beech woodlands are often open with a poorly developed shrub layer, Characteristic ground layer species may include various helleborines such as Cephalanthera damasonium, C. longifolia and C. rubra and sedges such as Carex alba, whilst in others, grasses like Sesleria caerlea or Melica uniflora may predominate, but in some of the more acidic examples, Luzula luzuloides is likely to dominate. There are also a number of endemic ground layer species. For example, in Carpathian beech woods endemics such as Dentaria glandulosa (Brassicaceae), Symphytum cordata (Boraginaceae) and the fern Polystichum braunii (Dryopteridaceae) may be encountered. Fine examples of primeaval beech woods can be found in the limestone Alps of lower Austria including the famous ‘Rothwald’ on the southeastern slopes of Dürrentein near Lunz. These range in altitude from about 940-1480 m. Here the canopy is dominated by Fagus sylvatica together with Acer pseudoplatanus, Picea abies, Ulmus glabra, and on the more acidic soils by Abies alba. Typical shrubs include Daphne mezereum, Lonicera alpigena and Rubus hirtus. At ground level the herb layer is very rich supporting possibly up to a 100 species of vascular plants. Examples include Adenostyles alliariae, Asplenium viridis, Campanula scheuchzeri, Cardamine trifolia, Cicerbita alpina, Denteria enneaphyllos, Euphorbia amygdaloides, Galium austriacum, Homogyne alpina, Lycopodium annotinum, Mycelis muralis, Paris quadrifolia, Phyteuma spicata, Prenanthes purpurea, Senecio fuchsii, Valeriana tripteris, Veratrum album and the central European endemic Helliborus niger (Ranunculaceae). -

23Rd International Workshop of the European Vegetation Survey
23rd International Workshop of the European Vegetation Survey Ljubljana 8–12 May 2014 Book of Abstracts 23rd International Workshop of the European Vegetation Survey – Book of Abstracts Editors: Andraž Čarni, Nina Juvan, Daniela Ribeiro Issued by: Jovan Hadži Institute of Biology ZRC SAZU Anton Melik Geographical Institute ZRC SAZU Represented by: Matjaž Kuntner, Drago Perko Technical editor: Iztok Sajko Published by: ZRC Publishing House Represented by: Oto Luthar Chief Editor: Aleš Pogačnik Printrun: 260 Ljubljana, May 2014 CIP - Kataložni zapis o publikaciji Narodna in univerzitetna knjižnica, Ljubljana 581.9(4)(082) EUROPEAN Vegetation Survey. International Workshop (23 ; 2014 ; Ljubljana) Book of abstracts / 23rd International Workshop of the European Vegetation Survey, Ljubljana 8-12 May 2014 ; [editors Andraž Čarni, Nina Juvan, Daniela Ribeiro]. - Ljubljana : ZRC Publishing House, 2014 ISBN 978-961-254-693-9 1. Čarni, Andraž 273600256 Contents Welcome ..............................................................................................................5 Organising Committee ..........................................................................................6 Scientific Committee ............................................................................................6 Program ................................................................................................................7 Poster presentation ............................................................................................14 Oral presentations -

SRGC BULB LOG DIARY---Pictures and Text © Ian Young
SRGC ----- Bulb Log Diary ----- ISSN 2514-6114 Pictures and text © Ian Young BULB LOG 25.....................17th June 2020 Meconopsis baileyi and Corydalis mucronipetala We planted out a number of Corydalis mucronipetala seedlings in this bed and now they are starting to seed. Earlier in the season the flowering colour in the New Bed Beside the Pond was provided by smaller early flowering bulbs, now these have completed their seasons growth as just a few Trillium hibbersonii and rivale are still to shed their seed, the larger plants such as the Meconopsis, Corydalis, make use of the same space. My interest in plants started when I was a boy in the wilderness of the Scottish mountains so it is not surprising that I encourage the garden to become wilder as I let nature take control with minimal intervention. I take great pleasure from wild moments like this combination of Celmisia, sprawling from where it was originally planted, being joined by a number of the plants that we allow to self-seed. The majority of these pictures were taken in a thick haar (mist) which along with some rain is bringing a welcome relief to the garden. I have been worried about the effect of stress, caused by our unusual long dry spring, on the plants we grow but now they all look refreshed with moisture hanging thick on their leaves and at their roots. We encourage all the bulbs to set seed as well as allowing their leaves to die down naturally so we can enjoy the colour change as they turn yellow on the ground, in contrast with the plants still in full fresh green growth.