Lithographx Documentation Release 1.2.0
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Postgis 3.1.0Alpha1 Manual I
PostGIS 3.1.0alpha1 Manual i PostGIS 3.1.0alpha1 Manual PostGIS 3.1.0alpha1 Manual ii Contents 1 Introdução 1 1.1 Comitê Diretor do Projeto . .1 1.2 Contribuidores Núclero Atuais . .2 1.3 Contribuidores Núclero Passado . .2 1.4 Outros Contribuidores . .2 2 Instalação do PostGIS 5 2.1 Versão Reduzida . .5 2.2 Compilando e instalando da fonte: detalhado . .5 2.2.1 Obtendo o Fonte . .6 2.2.2 Instalando pacotes requeridos . .6 2.2.3 Configuração . .7 2.2.4 Construindo . .9 2.2.5 Contruindo extensões PostGIS e implantado-as . .9 2.2.6 Testando . 11 2.2.7 Instalação . 20 2.3 Instalando e usando o padronizador de endereço . 21 2.3.1 Instalando Regex::Montar . 21 2.4 Instalando, Atualizando o Tiger Geocoder e carregando dados . 21 2.4.1 Tiger Geocoder ativando seu banco de dados PostGIS: Usando Extensão . 22 2.4.1.1 Convertendo uma Instalação Tiger Geocoder Regular para Modelo de Extensão . 24 2.4.2 Tiger Geocoder Ativando seu banco de dados PostGIS: Sem Utilizar Extensões . 24 2.4.3 Usando Padronizador de Endereço com Tiger Geocoder . 25 2.4.4 Carregando Dados Tiger . 25 2.4.5 Atualizando sua Instalação Tiger Geocoder . 26 2.5 Problemas comuns durante a instalação . 26 PostGIS 3.1.0alpha1 Manual iii 3 PostGIS Administration 28 3.1 Tuning your configuration for performance . 28 3.1.1 Startup . 28 3.1.2 Runtime . 29 3.2 Configuring raster support . 29 3.3 Creating spatial databases . 30 3.3.1 Spatially enable database using EXTENSION . -

Vefþjónustur SFR
Vefþjónustur SFR Vefþjónustur SÍ - SFR Efnisyfirlit Vefþjónustur SÍ - SFR Efnisyfirlit 1. Almennt 2. Slóðir 2.1. Skyggnir 2.1.1. Prófunarumhverfi Skyggnir 2.1.2. Raunumhverfi Skyggnir 2.2. TR/SÍ 2.2.1. Prófunarumhverfi TR/SÍ 2.2.2. Raunumhverfi TR/SÍ 3. Umslag : sfr 3.1. profun 3.2. stadasjuklings 3.3. vistaskjal 3.4. tryggingaskra 4. Stoðgögn 4.1. Villulisti 4.2. Staða sjúklings : tafla 4.3. Þjónustuflokkar sjúkrahúsa 4.4. Þjónustflokkar heilsugæslu (hér bætist oft nýtt við með nýjum sendendum) 4.5. TR-kóði: 5. SFR-soap köll 5.1. SFR-profun 5.2. SFR-stadasjuklings 5.3. SFR-vistaskjal 5.4. profun: 5.5. stadasjuklings: 5.6. vistaskjal: 1. Almennt Föll sem viðskiptavinir geta sent SÍ eru móttekin í gegnum SOAP-umslag. Umslag sfr Umslag fyrir upplýsingar tengdar ýmsum lækniskostnaði og útreikningi á komugjöldum. Upplýsingar sem fara á milli grunnkerfa SÍ og kerfa viðskiptavina SÍ. 2. Slóðir 2.1. Skyggnir Föll sem eru uppsett hjá Skyggni eru: profun stadasjuklings tryggingaskra Þau eru uppsett á eftirfarandi slóðum: 2.1.1. Prófunarumhverfi Skyggnir Prófunarumhverfi : https://pws.sjukra.is/sfr/sfr.svc Schema skilgreining : https://pws.sjukra.is/sfr/sfr.svc?wsdl 2.1.2. Raunumhverfi Skyggnir Raunumhverfi : https://ws.sjukra.is/sfr/sfr.svc Schema skilgreining : https://ws.sjukra.is/sfr/sfr.svc?wsdl 2.2. TR/SÍ Föllin sem eru uppsett hjá TR/SÍ eru: profun stadasjuklings vistaskjal Þau eru uppsett á eftirfarandi slóðum 2.2.1. Prófunarumhverfi TR/SÍ Prófunarumhverfi : https://huld.sjukra.is/p/sfr Schema skilgreining : https://huld.sjukra.is/p/sfr?wsdl 2.2.2. -

STOC-AGENT 講習会 (コンパイル編) 海岸港湾研究室(有川研究室) Installer
STOC-AGENT 講習会 (コンパイル編) 海岸港湾研究室(有川研究室) Installer 1) • MSMPI(Microsoft MPI v10.0 (Archived)) -msmpisdk.msi 前回配布分との変更点 -msmpisetup.exe 2) • gfortran Compiler(Mingw-w64) -mingw-w64-install.exe 3) • GNU MAKE -make-3.81.exe 4) • CADMAS-VR -CadmasVR_3.1.1_Setup_21050622.exe • CADMAS-MESH-MULTI 4) -CADMAS-MESH-MULTI-1.3.4-x64-setup.exe REFERED 1) https://www.microsoft.com/en-us/download/details.aspx?id=56727 2) http://mingw-w64.org/doku.php/download/mingw-builds 3) http://www.gnu.org/software/make/ http://gnuwin32.sourceforge.net/packages/make.htm 4) https://www.pari.go.jp/about/ MinGW-gfortran 1.以下のサイトからMingw-w64のダウンロードを行うため画面のSourceforgeをク リック. http://mingw-w64.org/doku.php/download/mingw-builds 2.MingW-W64-build を選択し、一番右図のような画面に移る. MinGW-gfortran 3. ダウンロードしたmingw-w64-installを実行インストールします. 基本的には変更なし 3. mingw-w64がインストールされていることを確認 MinGW-gfortran 5. コントロール パネル¥システムとセキュリティ¥システム¥システムの詳細設定 6. 環境変数を開き、ユーザーの環境変数,PATHを編集(PATHもしくはpathがなけれ ば新規で変数名にPATH,変数値に8.のアドレスを入力) 7. 環境変数名の編集→新規をクリック 8. gfortran.exeのあるフォルダのアドレスを入力(おそらくC:¥Program Files (x86) ¥mingw-w64¥i686-8.1.0-posix-dwarf-rt_v6-rev0¥mingw32¥bin) C:¥Program Files (x86)¥mingw-w64¥i686-8.1.0- posix-dwarf-rt_v6-rev0¥mingw32¥bin MinGW-gfortran 9.コマンドプロンプト(cmd)を開き,gfortran –v のコマンドを入力.以下 のような画面になれば環境設定完了(gfortranのPATHが通りました) GNU MAKE 1.以下のサイトからのmake.exeのダウンロードを行うため、画面のComplete packageのSetupをクリック. http://gnuwin32.sourceforge.net/packages/make.htm 2. make-3.8.1.exeを実行しインストールしてください. GnuWin32がインストールされていることを確認します. (おそらくC:¥Program Files (x86)¥) GNU MAKE 3. コントロール パネル¥システムとセキュリティ¥システム¥システムの詳細設定 4. 環境変数を開き、ユーザーの環境変数,PATHを編集(PATHもしくはpathがなけれ ば新規で変数名にPATH,変数値に6.のアドレスを入力) 5. 環境変数名の編集→新規をクリック 6. make.exeのあるフォルダのアドレスを入力 (おそらくC:¥Program Files (x86)¥GnuWin32¥bin) C:¥Program Files (x86)¥GnuWin32¥bin MSMPI 1.以下のサイトからMicrosoft MPI v10.0のダウンロードをクリック. https://www.microsoft.com/en-us/download/details.aspx?id=57467 2.msmpisdk.msi とmsmpisetup.exe をダウンロード. 3. -
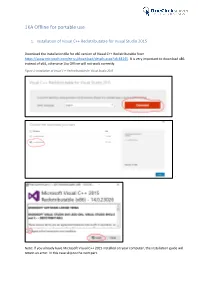
1KA Offline for Portable Use
1KA Offline for portable use 1. Installation of Visual C++ Redistributable for Visual Studio 2015 Download the installation file for x86 version of Visual C++ Redistributable from https://www.microsoft.com/en-us/download/details.aspx?id=48145. It is very important to download x86 instead of x64, otherwise 1ka Offline will not work correctly. Figure 1: Installation of Visual C++ Redistributable for Visual Studio 2015 Note: If you already have Microsoft Visual C++ 2015 installed on your computer, the installation guide will return an error. In this case skip to the next part. 2. Download 1KA Offline installation pack Download 1KA Offline from https://www.1ka.si/1ka-offline and unzip the file in the package to your hard drive (C:). First, click the button »Extract to« and then select Local Disk (C:). (Figure 2) Figure 2: Extracting of the installation package After the package is extracted, open the file »UwAmp« you have just created and start the application UwAmp by clicking on »UwAmp.exe«. (Figure 3). You must repeat this step every time you want to use 1KA offline (you can also create a shortcut on the desktop for easy access to the file). Figure 3: File UwAmp.exe 3. Windows users: Installation of additional libraries for data display If you are a Windows user, you must complete the following steps to install additional libraries that enable data display in 1KA Offline. The libraries can be accessed via: • Sed: http://gnuwin32.sourceforge.net/packages/sed.htm • Gawk: http://gnuwin32.sourceforge.net/packages/gawk.htm • Coreutils: http://gnuwin32.sourceforge.net/packages/coreutils.htm You can download the installation files by clicking »Setup« (as seen on figure 4). -

Building Microsoft Windows Versions of R and R Packages Under Intel Linux
BUILDING MICROSOFT WINDOWS VERSIONS OF R AND R PACKAGES UNDER INTEL LINUX Building Microsoft Windows Versions of R and R packages under Intel Linux A Package Developer’s Tool Linux R is in the search path, then by Jun Yan and A.J. Rossini make CrossCompileBuild will run all Makefile targets described up to the section, Disclaimer Building R Packages and Bundles. This assumes: (1) you have the Makefile in a directory RCrossBuild (empty ex- Cross-building R and R packages are documented in the R cept for the makefile), and (2) that ./RCrossBuild is your [1] source by the R Development Core Team, in files such as current working directory. After this, one should man- INSTALL, readme.package, and Makefile, under the directory ually set up the packages of interest for building, though src/gnuwin32/. This point was not clearly stated in an earlier the makefile will still help with many important steps. We version of this document [2] and has caused confusions. We describe this in detail in the section on Building R Packages apologize for the confusion and claim sole responsibility [3]. In and Bundles. addition, we clarify that the final credit should go to the R De- velopment Core Team. We intended to automate and illustrate those steps by presenting an explicit example, hoping that it Setting up the Build area might save people’s time. Follow them at your own risk. Puz- zles caused by this document should NOT be directed to the R We first create a separate directory for R cross-building, Development Core Team. -

Vefþjónustur SFR - Komureikningur
Vefþjónustur SFR - komureikningur Vefþjónustur SÍ - SFR Föll sem viðskiptavinir geta sent SÍ eru móttekin í gegnum SOAP-umslag. Umslag sfr Umslag fyrir upplýsingar tengdar ýmsum lækniskostnaði og útreikningi á komugjöldum. Upplýsingar sem fara á milli grunnkerfa SÍ og kerfa viðskiptavina SÍ. Prófunarumhverfi : https://huld.sjukra.is/p/sfr Schema skilgreining : https://huld.sjukra.is/p/sfr?wsdl Ath:. Tegund gilda eru annað hvort N = númer S=strengur D=Dagsetning. Tala innan sviga eftir tegund segir til um mögulega hámarksstærð þeirra. ( dæmi: N(10) er allt að 10 stafa tala). Dagsetningasvæði eru á forminu yyyy-mm-dd [ ] utan um tegundarskilgreiningu svæði merkir að svæði sé valkvætt. Efnisyfirlit Vefþjónustur SÍ - SFR Efnisyfirlit 1. Umslag : sfr 1.1. profun 1.2. stadasjuklings 1.3. vistaskjal 2. Schema: Reikningur komugjalda 3. Skýringar við xml-tög 4. Skema 5. Stoðgögn 5.1. Villulisti 5.2. Staða sjúklings : tafla 5.3. Þjónustuflokkar sjúkrahúsa 5.4. Þjónustflokkar heilsugæslu (hér bætist oft nýtt við með nýjum sendendum) 5.5. TR-kóði: 6. SFR-soap köll 6.1. SFR-profun 6.2. SFR-stadasjuklings 6.3. SFR-vistaskjal 6.4. profun: 6.5. stadasjuklings: 6.6. vistaskjal: 6.7. Skema: 1. Umslag : sfr Sjá: http://huld.sjukra.is:8887/sfr?wsdl (fá prófunarslóð staðfesta hjá SÍ) 1.1. profun Prófunarfall til að prófa hvort samskipti eru í lagi Heiti Tegund Skýring svæðis Inntak sendandi S(100) Einkenni raunverulegs sendanda ef verið er að senda gögn fyrir hönd einhvers annars annars autt/sleppt. starfsmadur S(50) Einkenni starfsmanns sendanda ef sendandi er útfylltur annars autt/sleppt. Úttak tokst N(1) 1 ef móttaka tókst, 0 annars villulysing S(1000) Lýsing á villu ef móttaka tókst ekki. -

Matthew Kendall
Stellaris Toolchain - Matthew Kendall http://www.matthewkendall.com/freesoftware/stellaris-to... Matthew Kendall Search this site Navigation Free Software > Home Stellaris Toolchain Contact Electronics Free Software Notes on how to set up the toolchain for development in C on Luminary Micro's Stellaris ARM Cortex M3 microcontrollers. Photos StellarisWare StellarisWare is Luminary Micro's name for the useful set of libraries that they supply. DriverLib is the Peripheral Driver Library for the on-chip peripherals (and appears to have previously been used as the name of the package as a whole). GrLib is the Graphics Library that provides a set of graphics primitives and a widget set for creating graphical user interfaces on boards that have a graphical display. There is also a bootloader, some (target) utilities and (host) tools. StellarisWare is supplied with evaluation kits and also downloadable from Luminary Micro. Download the version associated with the target board you have to get appropriate example programs. GCC, binutils, et al CodeSourcery says "CodeSourcery, in partnership with ARM, Ltd., develops improvements to the GNU Toolchain for ARM processors and provides regular, validated releases of the GNU Toolchain". CodeSourcery also develops proprietary software, principally startup code and various libraries for use on the target, and a debug "sprite" for use on the development host. This is all bundled together in a package named Sourcery G++ which is available in Personal and Professional editions. Ignore this and download the Lite edition which contains just the 1 de 5 14/05/13 20:45 Stellaris Toolchain - Matthew Kendall http://www.matthewkendall.com/freesoftware/stellaris-to.. -

Digital Forensics for Network, Internet, and Cloud Computing.Pdf
Syngress is an imprint of Elsevier. 30 Corporate Drive, Suite 400, Burlington, MA 01803, USA This book is printed on acid-free paper. © 2010 Elsevier Inc. All rights reserved. No part of this publication may be reproduced or transmitted in any form or by any means, electronic or mechanical, including photo- copying, recording, or any information storage and retrieval system, without permission in writing from the publisher. Details on how to seek permission, further information about the Publisher’s permissions policies and our arrangements with organizations such as the Copyright Clearance Center and the Copyright Licensing Agency, can be found at our Web site: www.elsevier.com/permissions. This book and the individual contributions contained in it are protected under copyright by the Publisher (other than as may be noted herein). Notices Knowledge and best practice in this field are constantly changing. As new research and experience broaden our understanding, changes in research methods, professional practices, or medical treatment may become necessary. Practitioners and researchers must always rely on their own experience and knowledge in evaluating and using any information, methods, compounds, or experiments described herein. In using such information or methods, they should be mindful of their own safety and the safety of others, including parties for whom they have a professional responsibility. To the fullest extent of the law, neither the Publisher nor the authors, contributors, or editors, assume any liability for any injury and/or damage to persons or property as a matter of products liability, negligence or otherwise, or from any use or operation of any methods, products, instructions, or ideas contained in the material herein. -

How to Set up Path to .Exe Gnuwin32 Binary Files in Windows XP / Vista / 2003 / 2008 (Setting PATH to Executables on Windows)
Walking in Light with Christ - Faith, Computing, Diary Articles & tips and tricks on GNU/Linux, FreeBSD, Windows, mobile phone articles, religious related texts http://www.pc-freak.net/blog How to set up Path to .exe GNUWin32 binary files in Windows XP / Vista / 2003 / 2008 (Setting PATH to executables on Windows) Author : admin I've been working on a servers running Windows 2003 and Windows 2008 these days. As I wanted to be more flexible on what I can do from the command line I decided to install GNUwin (provides port of GNU tools), most of which are common part of any Linux distribution). Having most of the command line flexibility on a Windows server is a great thing, so I would strongly recommend GNUWin to any Windows server adminsitrator out there. Actually it's a wonderful thing that most of the popular Linux tools can easily be installed and used on Windows for more check GnuWin32 on sourceforge One of the reasons I installed Gnuwin was my intention to use the good old Linux tail command to keep an eye interactive on the IIS server access log files, which by the way for IIS webserver are stored by default in C:\Windows\System32\LogFiles\W3SVC1\*.log I've managed to install the GNUWin following the install instructions, not with too much difficulties. The install takes a bit of time, cause many packs containing different parts of the GNUWin has to be fetched. To install I downloaded the GNUWin installer available from GNUWin32's website and instructed to extracted the files into C:\Program Files\Gnuwin Then I followed the install instructions suggestions, e.g.: C:\> cd c:\Program Files\GnuWin C:\Program Files\GnuWin> download.bat .. -

Tti9xe Software User Guide
TTI9XE SOFTWARE USER GUIDE SS11001-1 Jan 2011 Testtron Technologies Inc TTI9XE Software User Guide SS11001-1 Copyright 2008-2011 Testtron Technoligies Inc. All rights reserved. Testtron makes no warranty for the use of its products and assumes no responsibility for any errors which may appear in this document. Nor does it make a commitment to update the information contained herein. Testtron retains the right to make changes to these specifications at any time, without notice. Testtron products are not intended for use in medical, life saving or life sustaining applications. All product names referenced herein are trademarks of their respective companies. Revision History Version Date Description 1.0 Jan 2011 Initial Release Testtron Technologies Inc. 2 Jan 2011 TTI9XE Software User Guide SS11001-1 Table of Contents 1. Overview .......................................................................................................................................... 4 1.1 Scope ...................................................................................................................................... 4 1.2 References .............................................................................................................................. 4 1.3 Acronyms ................................................................................................................................ 4 2. Toolchain Components .................................................................................................................... 5 2.1 -

Index Images Download 2006 News Crack Serial Warez Full 12 Contact
index images download 2006 news crack serial warez full 12 contact about search spacer privacy 11 logo blog new 10 cgi-bin faq rss home img default 2005 products sitemap archives 1 09 links 01 08 06 2 07 login articles support 05 keygen article 04 03 help events archive 02 register en forum software downloads 3 security 13 category 4 content 14 main 15 press media templates services icons resources info profile 16 2004 18 docs contactus files features html 20 21 5 22 page 6 misc 19 partners 24 terms 2007 23 17 i 27 top 26 9 legal 30 banners xml 29 28 7 tools projects 25 0 user feed themes linux forums jobs business 8 video email books banner reviews view graphics research feedback pdf print ads modules 2003 company blank pub games copyright common site comments people aboutus product sports logos buttons english story image uploads 31 subscribe blogs atom gallery newsletter stats careers music pages publications technology calendar stories photos papers community data history arrow submit www s web library wiki header education go internet b in advertise spam a nav mail users Images members topics disclaimer store clear feeds c awards 2002 Default general pics dir signup solutions map News public doc de weblog index2 shop contacts fr homepage travel button pixel list viewtopic documents overview tips adclick contact_us movies wp-content catalog us p staff hardware wireless global screenshots apps online version directory mobile other advertising tech welcome admin t policy faqs link 2001 training releases space member static join health -

Ada User Journal
ADA Volume 23 USER Number 1 JOURNAL March 2002 Contents page Editorial Policy for Ada User Journal 2 Editorial 3 News 5 Conference Calendar 44 Forthcoming Events 51 Ada UK 2002 Sponsors 64 Ada-Europe Associate Members (National Ada Organizations) Inside Back Cover Ada User Journal Volume 23, Number 1, March 2002 2 Editorial Policy for Ada User Journal Publication Original Papers Commentaries Ada User Journal – The Journal for the Manuscripts should be submitted in We publish commentaries on Ada and international Ada Community – is accordance with the submission software engineering topics. These published by Ada-Europe. It appears guidelines (below). may represent the views either of four times a year, on the last days of individuals or of organisations. Such March, June, September and All original technical contributions are articles can be of any length – December. Copy date is the first of the submitted to refereeing by at least two inclusion is at the discretion of the month of publication. people. Names of referees will be kept Editor. confidential, but their comments will Opinions expressed within the Ada Aims be relayed to the authors at the discretion of the Editor. User Journal do not necessarily Ada User Journal aims to inform represent the views of the Editor, Ada- readers of developments in the Ada The first named author will receive a Europe or its directors. programming language and its use, complimentary copy of the issue of the general Ada-related software Journal in which their paper appears. Announcements and Reports engineering issues and Ada-related We are happy to publicise and report activities in Europe and other parts of By submitting a manuscript, authors grant Ada-Europe an unlimited licence on events that may be of interest to our the world.