Gene Expression Profiling of Olfactory Neuroblastoma Helps Identify
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

BLOOD RESEARCH September 2020 ARTICLE
VOLUME 55ㆍNUMBER 3 ORIGINAL BLOOD RESEARCH September 2020 ARTICLE Use of generic imatinib as first-line treatment in patients with chronic myeloid leukemia (CML): the GIMS (Glivec to Imatinib Switch) study Maria Gemelli1#, Elena Maria Elli2#, Chiara Elena3, Alessandra Iurlo4, Tamara Intermesoli5, Margherita Maffioli6, Ester Pungolino7, Maria Cristina Carraro8, Mariella D’Adda9, Francesca Lunghi10, Michela Anghileri11, Nicola Polverelli12, Marianna Rossi13, Mattia Bacciocchi14, Elisa Bono3, Cristina Bucelli4, Francesco Passamonti6, Laura Antolini15, Carlo Gambacorti-Passerini2,14 1Oncology Unit, San Gerardo Hospital, ASST-Monza, 2Hematology Division and Bone Marrow Unit, Ospedale San Gerardo, ASST- Monza, Monza, 3Division of Hematology, Fondazione IRCCS Policlinico San Matteo, Pavia, 4Hematology Division, Foundation IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan, 5Hematology and Bone Marrow Transplant Unit, ASST Papa Giovanni XXIII, Bergamo, 6Hematology Unit, Ospedale di Circolo, Varese, 7Division of Hematology, ASST Grande Ospedale Metropolitano Niguarda, Milano, 8Hematology and Transfusional Medicine Unit, ASST Fatebenefratelli Sacco, Milan, 9Department of Hematology, ASST Spedali Civili di Brescia, Brescia, 10Hematology and Bone Marrow Transplantation Unit, San Raffaele Scientific Institute, IRCCS Milano, 11Oncology Department, ASST Lecco, Lecco, 12Chair of Hematology, Unit of Blood Diseases and Stem Cell Transplantation, University of Brescia, ASST Spedali Civili di Brescia, Brescia, 13Department of Hematology, Cancer Center, IRCCS Humanitas Research Hospital/Humanitas University, Rozzano, 14Department of Medicine and Surgery, University of Milano-Bicocca, 15Center of Biostatistics for Clinical Epidemiology, Department of Medicine and Surgery, University of Milano-Bicocca, Monza, Italy p-ISSN 2287-979X / e-ISSN 2288-0011 Background https://doi.org/10.5045/br.2020.2020130 Generic formulations of imatinib mesylate have been introduced in Western Europe since Blood Res 2020;55:139-145. -
Eurostat: Recognized Research Entity
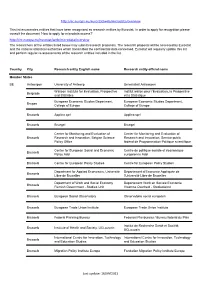
http://ec.europa.eu/eurostat/web/microdata/overview This list enumerates entities that have been recognised as research entities by Eurostat. In order to apply for recognition please consult the document 'How to apply for microdata access?' http://ec.europa.eu/eurostat/web/microdata/overview The researchers of the entities listed below may submit research proposals. The research proposal will be assessed by Eurostat and the national statistical authorities which transmitted the confidential data concerned. Eurostat will regularly update this list and perform regular re-assessments of the research entities included in the list. Country City Research entity English name Research entity official name Member States BE Antwerpen University of Antwerp Universiteit Antwerpen Walloon Institute for Evaluation, Prospective Institut wallon pour l'Evaluation, la Prospective Belgrade and Statistics et la Statistique European Economic Studies Department, European Economic Studies Department, Bruges College of Europe College of Europe Brussels Applica sprl Applica sprl Brussels Bruegel Bruegel Center for Monitoring and Evaluation of Center for Monitoring and Evaluation of Brussels Research and Innovation, Belgian Science Research and Innovation, Service public Policy Office fédéral de Programmation Politique scientifique Centre for European Social and Economic Centre de politique sociale et économique Brussels Policy Asbl européenne Asbl Brussels Centre for European Policy Studies Centre for European Policy Studies Department for Applied Economics, -

Demos / Posters
Demos / Posters The map of the rooms is available here DEMO SESSION 1 Tuesday, April 16 (10:30 - 16:00) Room: Corridoio Nizza Move-with-the-Sun or Move-with-the-Moon? Wide-area CloudNet Migrations Under Latency and Resource Constraints 1 / 14 Demos / Posters Johannes Grassler (TU Berlin & T-Labs, Germany); Stefan Schmid (T-Labs & TU Berlin, Germany); Anja Feldmann (TU-Berlin & Deutsche Telekom Laboratories, Germany) OPRSFEC: A Middleware of Packet Loss Recovery in Live Multicast Smart TV System Xiuyan Jiang (Fudan University, P.R. China); De jian Ye (Fudan University, P.R. China); Yiming Chen (Fudan University, P.R. China) Lossless Virtual Networks Daniel Crisan (ETHZ & IBM Research, Switzerland); Robert Birke (IBM Zurich Research Laboratory, Switzerland); Cyriel Minkenberg (IBM Research - Zurich, Switzerland); Mitchell Gusat (IBM Zurich research laboratory, Switzerland) Live Evaluation of Quality of Experience for Video Streaming and Web Browsing Louis Plissonneau (Orange Labs, France); Heng Cui (EURECOM, France); Ernst W Biersack (EURECOM, France) A User-centric Approach to Trust Management in Wi-Fi Networks Carlos Ballester (University of Geneva, Switzerland); Jean-Marc Seigneur (University of Geneva, Switzerland); Paolo Di Francesco (Level7, Italy); Valentin Moreno (FON, Spain); Rute C. Sofia (SITI, Universidade Lusófona, Portugal); Waldir Moreira (SITILabs, University Lusofona, Portugal); Alessandro Bogliolo (University of Urbino, Italy); Nuno Martins (Caixa Mágica Software, Portugal) User-centric Mobile Multimedia Service Delivery: -

Emidio Capriotti Phd CURRICULUM VITÆ
Emidio Capriotti PhD CURRICULUM VITÆ Name: Emidio Capriotti Nationality: Italian Date of birth: February, 1973 Place of birth: Roma, Italy Languages: Italian, English, Spanish Positions Oct 2019 Associate Professor: Department of Pharmacy and Biotechnology (FaBiT). University of Bologna, Bologna, Italy 2016-2019 Senior Assistant Professor (RTD type B): Department of Pharmacy and Biotechnology (FaBiT) and Department of Biological, Geological, and Environmental Sciences (BiGeA). University of Bologna, Bologna, Italy. 2015-2016 Junior Group Leader: Institute of Mathematical Modeling of Biological Systems, University of Düsseldorf, Düsseldorf, Germany 2012-2015 Assistant Professor: Division of Informatics, Department of Pathology, University of Alabama at Birmingham (UAB), Birmingham (AL), USA. 2011-2012 Marie-Curie IOF: Contracted Researcher at the Department of Mathematics and Computer Science, University of Balearic Islands (UIB), Palma de Mallorca, Spain. 2009-2011 Marie-Curie IOF: Postdoctoral Researcher at the Helix Group, Department of Bioengineering, Stanford University, Stanford (CA), USA. 2006-2009 Postdoctoral Researcher in the Structural Genomics Group at Department of Bioinformatics and Genetics, Prince Felipe Research Center (CIPF) Valencia, Spain. 2004-2006 Contract researcher at Department of Biology, University of Bologna, Bologna, Italy. 2001-2003 Ph.D student in Physical Sciences at University of Bologna, Bologna, Italy. Education Sep 2004 Master in Bioinformatics (first level) University of Bologna, Bologna (Italy). Jun 2004 Ph.D. in Physical Sciences University of Bologna, Bologna (Italy). Jul 1999 Laurea (B.S.) Degree in Physical Sciences, score 106/110 University of Bologna, Bologna (Italy). Visiting Jun 2012 – Jul 2012 Prof. Frederic Rousseau and Prof. Joost Schymkowitz, VIB Switch Laboratory, KU Leuven, Leuven (Belgium) May 2009 Prof. -

Curriculum Vitae Carlo Filippi
Curriculum Vitae Carlo Filippi Office address and URLs Department of Economics and Management - University of Brescia Contrada S. Chiara 50 - 25122 BRESCIA. Tel: +39 - 030 298 8576 E-mail: [email protected] OR group: http://or-brescia.unibs.it/ Scopus: https://www.scopus.com/authid/detail.uri?authorId=7006884051 Publons: https://publons.com/researcher/532100/carlo-filippi Google Scholar: http://scholar.google.it/citations?user=Wrwp_L0AAAAJ&hl=it CURRENT POSITION Associate Professor of Operations Research (MAT / 09) National Scientific Qualification, Full Professor, Operations Research. PROFESSIONAL EXPERIENCE 01/09/2006 – to the current date: Associate Professor of Operations Research (MAT / 09) University of Brescia - Department of Economics and Management, Brescia (Italy) Teaching: Currently Professor of Introduction to Logistics (6CFU, Laurea Master's Degree in Management), Supply Chain Management (6 CFU, Master's Degree in Management), Business Decision Analysis - AG (6 CFU, Bachelor's Degrees in Economics). Previously, professor and substitute teacher of various courses on logistics topics and decision support methods for master's degrees and on basic computer science topics and optimization for three-year degrees. Supervisor over 50 final reports for three-year and master's degrees in Economics, on topics related to logistics, optimization, information systems, electronic marketing. Research: Production of articles in international scientific journals and presentations at national and international sector conferences on topics -

Marginal Tumor Cysts As a Diagnostic MR Finding
Sinonasal Esthesioneuroblastoma with Intracranial Extension: Marginal Tumor Cysts as a Diagnostic MR Finding Peter M. Som, Mika Lidov, Margaret Brandwein, Peter Catalano, and Hugh F. Biller PURPOSE: To determine whether the MR finding of cysts along the intracranial margin of sinonasal esthesioneuroblastomas can be considered to suggest this tumor. METHODS: MR scans of 54 patients who had sinonasal lesions with intracranial extension were examined specifically for cysts along the intracranial margins of the lesions. RESULTS: Only 3 of the 54 patients had these cysts, and all 3 of these patients had esthesioneuroblastoma. Surgical pathologic findings of one speci men showed the cyst to be marginally located within the tumor. CONCLUSION: If cysts are seen on MR along the intracranial margin of a sinonasal mass, this finding highly suggests esthesia neuroblastoma. Index terms: Esthesioneuroblastoma; Nose, magnetic resonance AJNR Am J Neuroradio/15:1259-1262, Aug 1994 Radiologists often seem on a constant quest amelanotic melanomas, and embryonal rhab to give histologic diagnosis for disease seen on domyosarcomas ( 2). All of these neoplasms are sectional imaging studies. Rarely can this be undifferentiated, small-cell tumors, and like es accomplished. However, sometimes the pres thesioneuroblastoma can occur in the nasal ence of one or more imaging findings can allow fossa, with spread to the paranasal sinuses and the radiologist to offer a histologic diagnosis that anterior cranial fossa. Electron microscopy and has a very high degree of reliability. With this histochemical testing are required to establish a aim in mind, we examined the imaging studies definitive diagnosis (3). of 54 patients who had sinonasal masses with Initially, there was hope that radiologists extension into the anterior cranial fossa. -

(Brachytherapy) Education in Italy: Results of the INTERACTS Survey
Review Papers Review paper Current state of interventional radiotherapy (brachytherapy) education in Italy: results of the INTERACTS survey Luca Tagliaferri, MD, PhD1, György Kovács, MD, PhD2, Cynthia Aristei, MD3, Vitaliana De Sanctis, MD4, Fernando Barbera, MD5, Alessio Giuseppe Morganti, MD6, Calogero Casà, MD7, Bradley Rumwell Pieters, MD, PhD8, Elvio Russi, MD9, Lorenzo Livi, MD10, Renzo Corvò, MD11, Andrea Giovagnoni, MD12, Umberto Ricardi, MD13, Vincenzo Valentini, MD14,15, Stefano Maria Magrini, MD16 and the Directors of the Italian Radiation Oncology Schools** 1Chair of the Brachytherapy, Interventional Radiotherapy and IORT Study Group of the Italian Radiotherapy and Clinical Oncology Society (AIRO); Gemelli ART (Advanced Radiation Therapy), UOC Radioterapia, Fondazione Policlinico Universitario A. Gemelli IRCCS, Rome, Italy, 2INTERACTS (Interventional Radiotherapy Active Teaching School) Educational Program Director; Head, Interdisciplinary Brachytherapy Unit, University of Lübeck/UKSH CL, Lübeck, Germany, 3Past Chair of AIRO Brachytherapy study group; Head, Radiation Oncology Section, Department of Surgery and Biomedical Sciences, University of Perugia and Perugia General Hospital, Perugia, Italy, 4Deputy Chair of the Brachytherapy, Interventional Radiotherapy and IORT AIRO Study Group; Department of Radiation Oncology, Faculty of Medicina e Psicologia, Sant’Andrea Hospital, University of Rome “La Sapienza”, Rome, Italy, 5Board Member of the Brachytherapy, Interventional Radiotherapy and IORT AIRO Study Group; Head, Brachytherapy -

Dr Benedicta Marzinotto
CURRICULUM VITAE BENEDICTA MARZINOTTO Contact details: Rue de la Charité 33, B-1210 Brussels, Belgium [email protected] CURRENT EMPLOYMENT 2010- Research fellow, Bruegel, Brussels, Belgium 2005- Lecturer in Political Economy, Economics Department, University of Udine, Italy EDUCATION 2005 PhD in European Political Economy, London School of Economics (LSE), UK 2002 MPhil in European Political Economy, LSE, UK 1999 MSc in European Studies, LSE, UK RESEARCH INTERESTS European economic governance; political economy of fiscal adjustment; central banks and wage bargaining systems; varieties of capitalism; international trade and New Keynesian models. FELLOWSHIPS, HONOURS AND GRANTS 2005-10 Associate Fellow, International Economics, Chatham House, London, UK 2008 Visiting Scholar, University of Auckland, New Zealand 2007 Visiting Fellow, DGECFIN, European Commission, Brussels, Belgium 2005-07 Academic Collaboration – International Network Grant, Leverhulme Trust 2000-05 LSE Research Studentship 2004 Visiting Research Associate, Free University of Berlin, Germany 2004 UACES Research Scholarship 2002 Visiting Researcher, European University Institute, Fiesole, Italy TEACHING EXPERIENCE 2011- “Monetary Policy”, University of Udine, Italy 2009- “EU Macroeconomic Policy and Economic Governance”, College of Europe, Natolin Campus, Warsaw, Poland 2005- “Macroeconomics”, University of Udine, Italy 1 2006-08 “Government and Policies of the European Union”, MSc Mercosur and the EU in Comparative Perspective, Universidad Nacional de Cuyo, -

International Incoming Student's Guide
INTERNATIONAL INCOMING STUDENT’S GUIDE What you need to get the most out of Pavia and its university, and survive! UNIVERSITÀ DEGLI STUDI DI PAVIA Corso Strada Nuova 65, 27100 Pavia, Italy e-mail: [email protected] Tel. +39 0382 984021 Fax +39 0382 984695 Index 1. INTRODUCTION 9 1.1 THE INTERNATIONAL RELATIONS OFFICE AND WELCOME POINT 9 1.2 THE ITALIAN ACADEMIC SYSTEM 9 1.3 FACULTIES 10 1.4 PLACES YOU NEED FOR YOUR STUDIES 11 2. MEANS OF TRANSPORT 12 2.1 AIRPLANES AND AIRPORTS GETTING TO PAVIA 12 2.2 TRAINS AND STATIONS 12 2.2.1 TICKET BIGLIETTO 12 2.3 BUSES AND COACHES AUTOBUS E CORRIERE 13 3. BUREAUCRACY 15 3.1 LOST PASSPORT 15 4. UNIVERSITY OF PAVIA 16 4.1 SERVICES 16 4.1.1 CANTEENS MENSA 16 4.1.2 COMPUTER FACILITIES AND WI-FI CONNECTION 17 4.1.3 BOOKS LIBRI 17 4.1.4 LIBRARIES AND OPAC BIBLIOTECHE 17 4.1.5 BOOKSTORES LIBRERIE 18 4.2 UNIVERSITY LIFE VITA UNIVERSITARIA 19 4.2.1 C.U.S. UNIVERSITY SPORTS CENTRE 19 4.2.2 S.A.I.S.D. FACILITIES FOR DISABLED STUDENTS 19 4.2.3 STUDENT ASSOCIATIONS 19 4.2.4 MERCHANDISING 20 4.2.5 UCAMPUS 20 5. HEALTH 21 5.1 HEALTH INSURANCE ASSICURAZIONE SANITARIA 21 5.2 FIRST AID PRONTO SOCCORSO 21 5.3 PHARMACIES FARMACIA 22 5.4 MEDICINES MEDICINALI 23 6. FOOD AND FUN 25 6.1 SUPERMARKETS AND MARKET PLACES 25 6.2 BARS, PIZZERIAS AND RESTAURANTS 26 6.3 NIGHTLIFE - WHAT DOES PAVIA OFFER AT NIGHT? 27 7. -

Headmirror's ENT in a Nutshell Esthesioneuroblastoma Experts: Garret Choby, M.D. and Jamie Van Gompel, M.D. Presentation (0:28
Headmirror’s ENT in a Nutshell Esthesioneuroblastoma Experts: Garret Choby, M.D. and Jamie Van Gompel, M.D. Presentation (0:28) - Symptomatology o Nasal obstruction o Hyposmia or anosmia o Epistaxis o Headache, vision changes o Rare presentation cervical metastasis (5% patients) - Physical Examination o High nasal vault tumor (olfactory cleft) o Fleshy mass, moderately vascular o Thorough cranial nerve and neck examination - Epidemiology o Bimodal age distribution o SEER database middle to late age (40-60s) - Differential Diagnosis o Squamous cell carcinoma o Sinonasal undifferentiated carcinoma o Sinonasal neuroendocrine carcinoma o Lymphoma (imaging can look very similar) o Rhabdomyosarcoma o Ewing Sarcoma o Sinonasal mucosal melanoma Pathophysiology (2:20) - Theorized to arise from bipolar cell from nasal epithelium (upper 1.5 cm of the nasal vault) - Small blue round cell tumor or neuroendocrine tumor - Homer-Wright rosettes (1/3 of patients) - Flexner wintersteiner rosettes Workup (6:00) - Imaging o CT scan . Boney erosion at the skull base o MRI . Uniformly enhancing mass in the upper nasal vault with possible extension into the paranasal sinuses and intracranially . Dumbbell appearance is not reliable sign clincally o PET-CT scan . Especially for Kadish C-D lesions . Metastatic work up usually not performed for small lesions or Hyams grade 1-2 - Biopsy o Most important next step after imaging, especially in clinic - Grading System o Kadish Staging System . A: confined into the nasal cavity . B: extension into the paranasal sinuses . C: tumor beyond nasal cavity and paranasal sinus which can include cribiform plate, erosion of skull base, intracranial cavity or orbital invasion . D: cervical nodes or distant metastasis o Hyams Grading System . -

Midwifery Education Institutions in Italy Creation and Validation of Clinical Preceptors’ Assessment Tool: Students’ and Expert Midwives’ Views
Article Midwifery Education Institutions in Italy Creation and Validation of Clinical Preceptors’ Assessment Tool: Students’ and Expert Midwives’ Views Paola Agnese Mauri 1,2,* , Ivan Cortinovis 1 , Norma Nilde Guerrini Contini 2 and Marta Soldi 2 1 Department of Clinical Sciences and Community Health, Università degli Studi di Milan, via Manfredo Fanti 6, 20122 Milan, Italy; [email protected] 2 Unit of Mother Child and Newborn Health, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, via Manfredo Fanti 6, 20122 Milan, Italy; [email protected] (N.N.G.C.); [email protected] (M.S.) * Correspondence: [email protected]; Tel./Fax: +39-025-503-8599 Received: 20 November 2020; Accepted: 14 December 2020; Published: 16 December 2020 Abstract: Background: The aim of the study is to create and validate a midwifery preceptor’s evaluation form to be used by midwifery students. The International Confederation of Midwives recommends that clinical placements need to be supervised by a preceptor in order to be efficient for students who, in this way, gain competence and proper practice within the midwifery practical area. Methods: This is an observational multi-center transversal study and leads to the validation of an evaluation questionnaire. Methodically, the following steps were followed: literature review, focus group with midwifery students, meeting between expert midwives, creation of the preceptor’s assessment form, filling in of the forms by midwifery students and expert midwives, and validation of the form. The study was carried out in eight Italian universities and included eighty-eight midwifery students and eight midwives. Results and Conclusion: A midwifery preceptor’s assessment questionnaire was created made up of four attribute areas which, as a total, included 33 items. -

Endonasal Endoscopic Approach for Skull Base Neurofibroma: Is It Viable? 1Ashok Gupta, 2MK Tiwari, 3Neha Chauhan, 4Navjot Kaur, 5Vaiphei Kim
AIJCR Ashok Gupta et al 10.5005/jp-journals-10013-1237 CASE REPORT Endonasal Endoscopic Approach for Skull Base Neurofibroma: Is It Viable? 1Ashok Gupta, 2MK Tiwari, 3Neha Chauhan, 4Navjot Kaur, 5Vaiphei Kim ABSTRACT CASE REPORT Anterior skull base and sinonasal schwannomas are rare A 25 years female presented to outpatient services of entities. Earlier, safest way to remove these tumors, with best Department of Otorhinolaryngology—Head and Neck success rates, was open craniofacial surgery but recently with Surgery, Postgraduate Institute of Medical Education introduction of endoscopic endonasal approach; complete and Research, Chandigarh with complaints of left sided resection of these rare entities is possible. We present one facial pain since 20 years. The pain was intermittent and such case of skull base neurofibroma which was resected in entirety via a purely endoscopic endonasal approach. episodic. The episode lasted for 15 to 20 minutes and was mild to moderate in intensity. It was more on the frontal Keywords: Endonasal, Endoscopic, Schwannoma, Skull base. region on left side and extended to the left eye and face. It How to cite this article: Gupta A, Tiwari MK, Chauhan N, was relieved with medication in the form of pain killers. Kaur N, Kim V. Endonasal Endoscopic Approach for Skull Base There was no history of nasal discharge, altered smell, Neurofibroma: Is It Viable?. Clin Rhinol An Int J 2015;8(2):72-75. decreased or double vision, seizures, fever, neck rigidity Source of support: Nil of any focal neurological deficit. There was no significant past medical or surgical, family or personal history. Conflict of interest: None On examination, her ENT examination was grossly normal.