Genetic Diversity Analysis of Crab Spider (Araneae: Thomisidae) Based on RAPD-PCR
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Checklist of the Non -Acarine Arachnids
Original Research A CHECKLIST OF THE NON -A C A RINE A R A CHNIDS (CHELICER A T A : AR A CHNID A ) OF THE DE HOOP NA TURE RESERVE , WESTERN CA PE PROVINCE , SOUTH AFRIC A Authors: ABSTRACT Charles R. Haddad1 As part of the South African National Survey of Arachnida (SANSA) in conserved areas, arachnids Ansie S. Dippenaar- were collected in the De Hoop Nature Reserve in the Western Cape Province, South Africa. The Schoeman2 survey was carried out between 1999 and 2007, and consisted of five intensive surveys between Affiliations: two and 12 days in duration. Arachnids were sampled in five broad habitat types, namely fynbos, 1Department of Zoology & wetlands, i.e. De Hoop Vlei, Eucalyptus plantations at Potberg and Cupido’s Kraal, coastal dunes Entomology University of near Koppie Alleen and the intertidal zone at Koppie Alleen. A total of 274 species representing the Free State, five orders, 65 families and 191 determined genera were collected, of which spiders (Araneae) South Africa were the dominant taxon (252 spp., 174 genera, 53 families). The most species rich families collected were the Salticidae (32 spp.), Thomisidae (26 spp.), Gnaphosidae (21 spp.), Araneidae (18 2 Biosystematics: spp.), Theridiidae (16 spp.) and Corinnidae (15 spp.). Notes are provided on the most commonly Arachnology collected arachnids in each habitat. ARC - Plant Protection Research Institute Conservation implications: This study provides valuable baseline data on arachnids conserved South Africa in De Hoop Nature Reserve, which can be used for future assessments of habitat transformation, 2Department of Zoology & alien invasive species and climate change on arachnid biodiversity. -

Supraspecific Names in Spider Systematic and Their Nomenclatural Problems
Arachnologische Mitteilungen / Arachnology Letters 55: 42-45 Karlsruhe, April 2018 Supraspecific names in spider systematic and their nomenclatural problems Yuri M. Marusik doi: 10.30963/aramit5507 Abstract. Three different types of the names used in spider systematics are recognized and discussed: 1) typified taxonomic names, 2) non-typified taxonomic names, and 3) non-taxonomic names. Typified names are those from genus to superfamily group names; they are regulated by the ICZN. Non-typified names are used for taxonomic groups higher than superfamilies (e.g., Haplogynae, Mesothelae, etc.); they are not regulated by the ICZN but have an authorship, a fixed year of publication and are incorporated in a hierarchical classification. Non-taxonomic names are not regulated by any formal rules, unranked, have no authorship or description, and are non-typified. Some difficulties connected with the non-typified names in spider systematics are briefly discussed. Senior synonyms of some non-typified and non-taxonomic names are discussed, and suggestions are given on how to deal with the non-typified names lacking senior synonyms. Keywords: clade name, non-typified name, typified name. Zusammenfassung. Supraspezifische Namen in der Spinnensystematik und ihre nomenklatorischen Probleme. Drei verschie- dene Namenstypen in der Spinnensystematik werden diskutiert: 1) typisierte taxonomische Namen, 2) nicht-typisierte taxonomische Namen sowie 3) nicht-taxonomische Namen. Typisierte Namen reichen von Gattungen bis zu Überfamilien und sind durch die ICZN reguliert. Nicht-typisierte Namen werden für taxonomische Einheiten oberhalb von Überfamilien verwendet (z. B. Haplogynae, Meso- thelae), sind nicht durch die ICZN reguliert, haben aber Autoren, ein Erstbeschreibungsjahr und werden in hierarchischen Klassifikatio- nen verwendet. -

The Spiders Diversity from Different Habitats Around Biosciences, Vallabh Vidyanagar
International Journal of Science and Research (IJSR) ISSN (Online): 2319-7064 Index Copernicus Value (2013): 6.14 | Impact Factor (2014): 5.611 The Spiders Diversity from Different Habitats around Biosciences, Vallabh Vidyanagar B. M. Parmar Zoology Department, Sheth M.N. Science College, Patan, Gujarat-384 265 Abstract: A preliminary study of spiders was carried out in June 2012 to July 2013 from five sites around Biosciences, Vallabh Vidyanagar. As the results of collected spiders, total 90 species belonging to 66 genera spread over 24 families are recorded from five sites of Vallabhvidyanagar. The dominant family Araneidae had the highest number of species (18); followed by Salticidae (13), Thomisidae (10) and Tetragnathidae (7), Oxyopidae (5). Most of the other families had less than 5 species. This small region has detected more than 5% of Indian spiders. Keywords: Spiders, diversity, Anand, Gujarat 1. Introduction spiders with 12 species (13.33%) and irregular web builder 12 species (13.33%), Ambusher spiders with 11 (12.22%) Spiders of Gujarat from all regions have been studied earlier species each. The funnel web spiders with 7 species (7.77%) by several researchers; viz. Patel, B. H. [16], Patel, B. H. and and foliage hunter/ runner spiders with 3 species (3.33%). R.V.Vyas. [17], Manju Saliwal et. al., [6], Nikunj Bhatt, [10], Patel et. al., [18], Vachhani et. al., [26], Parmar Bharat Table 1: Sites Descriptions N. et. al., [15], Parmar, B.M. and K.B.Patel [12], Parmar, No. Site Geographic Habitat description B.M. et. al., [13], Parmar, B.M. and A.V.R.L.N. -

SHORT COMMUNICATION a Vertebrate-Eating Jumping Spider
2017. Journal of Arachnology 45:238–241 SHORT COMMUNICATION A vertebrate-eating jumping spider (Araneae: Salticidae) from Florida, USA Martin Nyffeler1, G. B. Edwards2 and Kenneth L. Krysko3: 1Section of Conservation Biology, Department of Environmental Sciences, University of Basel, CH-4056, Basel, Switzerland; E-mail: [email protected]; 2Curator Emeritus: Arachnida & Myriapoda, Florida State Collection of Arthropods, Gainesville, FL 32608, USA; 3Division of Herpetology, Florida Museum of Natural History, University of Florida, Gainesville, FL 32611, USA Abstract. The salticid spider Phidippus regius C.L. Koch, 1846 is documented preying on small frogs (Hyla spp., Osteopilus septentrionalis) and lizards (Anolis carolinensis and Anolis sagrei) in Florida, USA. Female as well as male P. regius were engaged in feeding on this type of vertebrate prey. A total of eight incidents of P. regius devouring vertebrates have been witnessed in seven Florida counties. Furthermore, we report an incident of a large unidentified Phidippus sp. (possibly P. bidentatus F. O. Pickard-Cambridge, 1901) preying on an immature anole lizard in Costa Rica. P. regius, otherwise known to feed almost exclusively on insects and spiders, is one of the world’s largest salticid spiders reaching a maximum recorded body length of 2.2 cm. Most other salticid spiders appear to be too small in body size to overcome vertebrate prey. Vertebrate predation by salticid spiders has not been previously documented in the scientific literature. Together with Salticidae, spiders from 27 of 114 families (24%) are currently known to occasionally consume vertebrate prey. Keywords: Generalist predators, predation, prey, Dactyloidae, Hylidae, Southeastern USA With .5,900 described species, the jumping spider family (Salt- observations. -
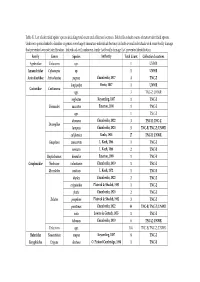
Table S1. List of Identified Spider Species Including Total Count and Collection Locations. Bold Cells Include Counts of Mature Identified Species
Table S1. List of identified spider species including total count and collection locations. Bold cells include counts of mature identified species. Unknown species linked to families or genera were largely immature individuals but may include several individuals with some bodily damage that prevented accurate identification. Individuals with unknown family had bodily damage that prevented identification. Family Genus Species Authority Total Count Collection Locations Agelenidae Unknown spp. 1 UNWR Amaurobiidae Cybaeopsis sp. 1 UNWR Antrodiaetidae Antrodiaetus pugnax Chamberlin, 1917 4 TNC-Z longipalpa Hentz, 1847 1 UNWR Corinnidae Castianeira spp. 3 TNC-Z; UNWR neglectus Keyserling, 1887 1 TNC-Z Drassodes saccatus Emerton, 1890 1 TNC-Z spp. 1 TNC-Z dromeus Chamberlin, 1922 3 TNC-B; TNC-Z Drassyllus lamprus Chamberlin, 1920 3 TNC-B; TNC-Z; UNWR californica Banks, 1904 17 TNC-B; UNWR Gnaphosa muscorum L. Koch, 1866 1 TNC-Z sericata L. Koch, 1866 2 TNC-B Haplodrassus hiemalis Emerton, 1909 1 TNC-B Gnaphosidae Nodocion voluntaries Chamberlin, 1919 1 TNC-Z Urozelotes rusticus L. Koch, 1872 1 TNC-B duplex Chamberlin, 1922 2 TNC-Z exiguioides Platnick & Shadab, 1983 1 TNC-Z fratis Chamberlin, 1920 2 TNC-Z Zelotes josephine Platnick & Shadab, 1983 3 TNC-Z puritanus Chamberlin, 1922 44 TNC-B; TNC-Z; UNWR sula Lowrie & Gertsch, 1955 1 TNC-Z tubuous Chamberlin, 1919 6 TNC-Z; UNWR Unknown spp. 184 TNC-B; TNC-Z; UNWR Hahniidae Neoantistea magna Keyserling, 1887 8 TNC-Z Linyphiidae Erigone dentosa O. Pickard-Cambridge, 1894 1 TNC-B spp. 1 TNC-Z Unknown spp. 13 TNC-Z; UNWR mccooki Montgomery, 1904 95 TNC-B; TNC-Z; UNWR Schizocosa minnesotensis Gertsch, 1934 25 TNC-B Lycosidae spp. -

Phylogeny of Entelegyne Spiders: Affinities of the Family Penestomidae
Molecular Phylogenetics and Evolution 55 (2010) 786–804 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Phylogeny of entelegyne spiders: Affinities of the family Penestomidae (NEW RANK), generic phylogeny of Eresidae, and asymmetric rates of change in spinning organ evolution (Araneae, Araneoidea, Entelegynae) Jeremy A. Miller a,b,*, Anthea Carmichael a, Martín J. Ramírez c, Joseph C. Spagna d, Charles R. Haddad e, Milan Rˇezácˇ f, Jes Johannesen g, Jirˇí Král h, Xin-Ping Wang i, Charles E. Griswold a a Department of Entomology, California Academy of Sciences, 55 Music Concourse Drive, Golden Gate Park, San Francisco, CA 94118, USA b Department of Terrestrial Zoology, Nationaal Natuurhistorisch Museum Naturalis, Postbus 9517 2300 RA Leiden, The Netherlands c Museo Argentino de Ciencias Naturales – CONICET, Av. Angel Gallardo 470, C1405DJR Buenos Aires, Argentina d William Paterson University of New Jersey, 300 Pompton Rd., Wayne, NJ 07470, USA e Department of Zoology & Entomology, University of the Free State, P.O. Box 339, Bloemfontein 9300, South Africa f Crop Research Institute, Drnovská 507, CZ-161 06, Prague 6-Ruzyneˇ, Czech Republic g Institut für Zoologie, Abt V Ökologie, Universität Mainz, Saarstraße 21, D-55099, Mainz, Germany h Laboratory of Arachnid Cytogenetics, Department of Genetics and Microbiology, Faculty of Science, Charles University in Prague, Prague, Czech Republic i College of Life Sciences, Hebei University, Baoding 071002, China article info abstract Article history: Penestomine spiders were first described from females only and placed in the family Eresidae. Discovery Received 20 April 2009 of the male decades later brought surprises, especially in the morphology of the male pedipalp, which Revised 17 February 2010 features (among other things) a retrolateral tibial apophysis (RTA). -

SA Spider Checklist
REVIEW ZOOS' PRINT JOURNAL 22(2): 2551-2597 CHECKLIST OF SPIDERS (ARACHNIDA: ARANEAE) OF SOUTH ASIA INCLUDING THE 2006 UPDATE OF INDIAN SPIDER CHECKLIST Manju Siliwal 1 and Sanjay Molur 2,3 1,2 Wildlife Information & Liaison Development (WILD) Society, 3 Zoo Outreach Organisation (ZOO) 29-1, Bharathi Colony, Peelamedu, Coimbatore, Tamil Nadu 641004, India Email: 1 [email protected]; 3 [email protected] ABSTRACT Thesaurus, (Vol. 1) in 1734 (Smith, 2001). Most of the spiders After one year since publication of the Indian Checklist, this is described during the British period from South Asia were by an attempt to provide a comprehensive checklist of spiders of foreigners based on the specimens deposited in different South Asia with eight countries - Afghanistan, Bangladesh, Bhutan, India, Maldives, Nepal, Pakistan and Sri Lanka. The European Museums. Indian checklist is also updated for 2006. The South Asian While the Indian checklist (Siliwal et al., 2005) is more spider list is also compiled following The World Spider Catalog accurate, the South Asian spider checklist is not critically by Platnick and other peer-reviewed publications since the last scrutinized due to lack of complete literature, but it gives an update. In total, 2299 species of spiders in 67 families have overview of species found in various South Asian countries, been reported from South Asia. There are 39 species included in this regions checklist that are not listed in the World Catalog gives the endemism of species and forms a basis for careful of Spiders. Taxonomic verification is recommended for 51 species. and participatory work by arachnologists in the region. -

Araneae : Philodromidae and Thomisidae, by Charles D
The Journal of Arachnology 8 : 95 Muchmore, W . B. 1969a. The pseudoscorpion genus Neochthonius Chamberlin (Arachnida , Chelonethida, Chthoniidae) with description of a cavernicolous species, Amer. Midl . Nat . 81 :387-394 . Muchmore, W. B . 1969b . New species and records of cavernicolous pseudoscorpions of the genu s Microcreagris (Arachnida, Chelonethida, Neobisiidae, Ideobisiinae) . Amer . Mus. Novitate s 2392:1-21 . Muchmore, W. B . 1976 . New species of Apochthonius, mainly from caves in central and eastern United States (Pseudoscirpionida, Chthoniidae) . Proc. Biol . Soc . Washington 89 :67-80 . Muchmore, W . B. and E. M . Benedict . 1976 . Redescription of Apochthonius moestus (Banks), type of the genus Apochthonius Chamberlin (Pseudoscirpionida, Chthoniidae) . J. New York Entomol. Soc. 84 :67-74 . Schuster, R . O . 1966 . New species of Apochthonius from western North America . Pan-Pacifi c Entomol . 42 :178-183 . Manuscript received March 1979, revised May 1979. BOOK REVIEW The Crab Spiders of Canada and Alaska : Araneae : Philodromidae and Thomisidae, by Charles D . Dondale and James H. Redner, 1978 . Part 5 of The Insects and Arachnida o f Canada, Publication 1663, pp . 1-255, 725 figs., 66 maps . Available from Printing and Publishing Supply and Services Canada, Hull, Quebec, Canada K1A OS9, Canada . Canada : $7 .50 ; other countries : $9.00. This handsome, soft-bound volume, dealing with the spider families Philodromidae an d Thomisidae from north of the United States, is a striking contribution to Canadia n arachnology . About half of the taxa of all temperate North America (110 of some 22 3 species) occur in Canada . The authors have drawn much of the data from their many papers on these spiders, but this has been materially supplemented by new appraisals , illustrations, and distribution information . -

L:\PM IJA Vol.3.Pmd
© Indian Society of Arachnology ISSN 2278 - 1587 SPIDER FAUNA OF RADHANAGARI WILDLIFE SANC- TUARY, CHANDOLI NATIONAL PARK AND KOYNA WILDLIFE SANCTUARY Suvarna More and Vijay Sawant* P. V. P. Mahavidyalaya, Kavathe Mahankal, Sangli. *Former Professor and Head, Department of Zoology, Shivaji University, Kolhapur. ABSTRACT Diversity of spiders from Radhanagari Wildlife Sanctuary, Chandoli Na- tional Park and Koyna Wildlife Sanctuary in Western Ghats is studied for the first time. A total of 247 species belonging to 119 genera and 28 families are recorded from the study area during 2010-2012 with a dominance of Araneid, Salticid and Lycosid spiders. Key words: Spider diversity, Western Ghats INTRODUCTION Spiders comprise one of the largest (5-6th) orders of animals. The spider fauna of India has never been studied in its entirety despite of contributions by many arachnologists since Stoliczka (1869). The pioneering contribution on the taxonomy of Indian spiders is that of European arachnologist Stoliczka (1869). Review of available literature reveals that the earliest contribution by Blackwall (1867); Karsch (1873); Simon (1887); Thorell (1895) and Pocock (1900) were the pioneer workers of Indian spiders. They described many species from India. Tikader (1980, 1982), Tikader, and Malhotra (1980a,b) described spiders from India. Tikader (1980) compiled a book on Thomisid spiders of India, comprising two subfamilies, 25 genera and 115 species. Of these, 23 species were new to science. Descriptions, illustrations and distributions of all species were given. Keys to the subfamilies, genera, and species were provided. Tikader and Biswas (1981) studied 15 families, 47 genera and 99 species from Calcutta and surrounding areas with illustrations and descriptions. -

Diversity of Common Garden and House Spider in Tinsukia District, Assam Has Been Undertaken
Journal of Entomology and Zoology Studies 2019; 7(4): 1432-1439 E-ISSN: 2320-7078 P-ISSN: 2349-6800 Diversity of common garden and house spider in JEZS 2019; 7(4): 1432-1439 © 2019 JEZS Tinsukia district Received: 01-05-2019 Accepted: 05-06-2019 Achal Kumari Pandit Achal Kumari Pandit Graduated from Department of Zoology Digboi College, Assam, Abstract India A study on the diversity of spider fauna inside the Garden and House in Tinsukia district, Assam. This was studied from September 2015 to July 2019. A total of 18 family, 52 genus and 80 species were recorded. Araneidae is the most dominant family among all followed by the silicide family. The main aim of this study is to bring to known the species which is generally observed by the humans in this area. Beside seasonal variation in species is higher in summer season as compared to winter. Also many species were observed each year in same season repeatedly during the study period, further maximum number of species is seen in vegetation type of habitat. Keywords: Spider, diversity, Tinsukia, seasonal, habitat 1. Introduction As one of the most widely recognized group of Arthropods, Spiders are widespread in distribution except for a few niches, such as Arctic and Antarctic. Almost every plant has its spider fauna, as do dead leaves, on the forest floor and on the trees. They may be found at varied locations, such as under bark, beneath stones, below the fallen logs, among foliage, [23] house dwellings, grass, leaves, underground, burrows etc. (Pai IK., 2018) . Their success is reflected by the fact that, on our planet, there are about 48,358 species recorded till now according to World Spider Catalog. -

Arachnologische Arachnology
Arachnologische Gesellschaft E u Arachnology 2015 o 24.-28.8.2015 Brno, p Czech Republic e www.european-arachnology.org a n Arachnologische Mitteilungen Arachnology Letters Heft / Volume 51 Karlsruhe, April 2016 ISSN 1018-4171 (Druck), 2199-7233 (Online) www.AraGes.de/aramit Arachnologische Mitteilungen veröffentlichen Arbeiten zur Faunistik, Ökologie und Taxonomie von Spinnentieren (außer Acari). Publi- ziert werden Artikel in Deutsch oder Englisch nach Begutachtung, online und gedruckt. Mitgliedschaft in der Arachnologischen Gesellschaft beinhaltet den Bezug der Hefte. Autoren zahlen keine Druckgebühren. Inhalte werden unter der freien internationalen Lizenz Creative Commons 4.0 veröffentlicht. Arachnology Logo: P. Jäger, K. Rehbinder Letters Publiziert von / Published by is a peer-reviewed, open-access, online and print, rapidly produced journal focusing on faunistics, ecology Arachnologische and taxonomy of Arachnida (excl. Acari). German and English manuscripts are equally welcome. Members Gesellschaft e.V. of Arachnologische Gesellschaft receive the printed issues. There are no page charges. URL: http://www.AraGes.de Arachnology Letters is licensed under a Creative Commons Attribution 4.0 International License. Autorenhinweise / Author guidelines www.AraGes.de/aramit/ Schriftleitung / Editors Theo Blick, Senckenberg Research Institute, Senckenberganlage 25, D-60325 Frankfurt/M. and Callistus, Gemeinschaft für Zoologische & Ökologische Untersuchungen, D-95503 Hummeltal; E-Mail: [email protected], [email protected] Sascha -

Programme and Abstracts European Congress of Arachnology - Brno 2 of Arachnology Congress European Th 2 9
Sponsors: 5 1 0 2 Programme and Abstracts European Congress of Arachnology - Brno of Arachnology Congress European th 9 2 Programme and Abstracts 29th European Congress of Arachnology Organized by Masaryk University and the Czech Arachnological Society 24 –28 August, 2015 Brno, Czech Republic Brno, 2015 Edited by Stano Pekár, Šárka Mašová English editor: L. Brian Patrick Design: Atelier S - design studio Preface Welcome to the 29th European Congress of Arachnology! This congress is jointly organised by Masaryk University and the Czech Arachnological Society. Altogether 173 participants from all over the world (from 42 countries) registered. This book contains the programme and the abstracts of four plenary talks, 66 oral presentations, and 81 poster presentations, of which 64 are given by students. The abstracts of talks are arranged in alphabetical order by presenting author (underlined). Each abstract includes information about the type of presentation (oral, poster) and whether it is a student presentation. The list of posters is arranged by topics. We wish all participants a joyful stay in Brno. On behalf of the Organising Committee Stano Pekár Organising Committee Stano Pekár, Masaryk University, Brno Jana Niedobová, Mendel University, Brno Vladimír Hula, Mendel University, Brno Yuri Marusik, Russian Academy of Science, Russia Helpers P. Dolejš, M. Forman, L. Havlová, P. Just, O. Košulič, T. Krejčí, E. Líznarová, O. Machač, Š. Mašová, R. Michalko, L. Sentenská, R. Šich, Z. Škopek Secretariat TA-Service Honorary committee Jan Buchar,