Genetic Timeline
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Glossary - Cellbiology
1 Glossary - Cellbiology Blotting: (Blot Analysis) Widely used biochemical technique for detecting the presence of specific macromolecules (proteins, mRNAs, or DNA sequences) in a mixture. A sample first is separated on an agarose or polyacrylamide gel usually under denaturing conditions; the separated components are transferred (blotting) to a nitrocellulose sheet, which is exposed to a radiolabeled molecule that specifically binds to the macromolecule of interest, and then subjected to autoradiography. Northern B.: mRNAs are detected with a complementary DNA; Southern B.: DNA restriction fragments are detected with complementary nucleotide sequences; Western B.: Proteins are detected by specific antibodies. Cell: The fundamental unit of living organisms. Cells are bounded by a lipid-containing plasma membrane, containing the central nucleus, and the cytoplasm. Cells are generally capable of independent reproduction. More complex cells like Eukaryotes have various compartments (organelles) where special tasks essential for the survival of the cell take place. Cytoplasm: Viscous contents of a cell that are contained within the plasma membrane but, in eukaryotic cells, outside the nucleus. The part of the cytoplasm not contained in any organelle is called the Cytosol. Cytoskeleton: (Gk. ) Three dimensional network of fibrous elements, allowing precisely regulated movements of cell parts, transport organelles, and help to maintain a cell’s shape. • Actin filament: (Microfilaments) Ubiquitous eukaryotic cytoskeletal proteins (one end is attached to the cell-cortex) of two “twisted“ actin monomers; are important in the structural support and movement of cells. Each actin filament (F-actin) consists of two strands of globular subunits (G-Actin) wrapped around each other to form a polarized unit (high ionic cytoplasm lead to the formation of AF, whereas low ion-concentration disassembles AF). -

(APOCI, -C2, and -E and LDLR) and the Genes C3, PEPD, and GPI (Whole-Arm Translocation/Somatic Cell Hybrids/Genomic Clones/Gene Family/Atherosclerosis) A
Proc. Natl. Acad. Sci. USA Vol. 83, pp. 3929-3933, June 1986 Genetics Regional mapping of human chromosome 19: Organization of genes for plasma lipid transport (APOCI, -C2, and -E and LDLR) and the genes C3, PEPD, and GPI (whole-arm translocation/somatic cell hybrids/genomic clones/gene family/atherosclerosis) A. J. LUSIS*t, C. HEINZMANN*, R. S. SPARKES*, J. SCOTTt, T. J. KNOTTt, R. GELLER§, M. C. SPARKES*, AND T. MOHANDAS§ *Departments of Medicine and Microbiology, University of California School of Medicine, Center for the Health Sciences, Los Angeles, CA 90024; tMolecular Medicine, Medical Research Council Clinical Research Centre, Harrow, Middlesex HA1 3UJ, United Kingdom; and §Department of Pediatrics, Harbor Medical Center, Torrance, CA 90509 Communicated by Richard E. Dickerson, February 6, 1986 ABSTRACT We report the regional mapping of human from defects in the expression of the low density lipoprotein chromosome 19 genes for three apolipoproteins and a lipopro- (LDL) receptor and is strongly correlated with atheroscle- tein receptor as well as genes for three other markers. The rosis (15). Another relatively common dyslipoproteinemia, regional mapping was made possible by the use of a reciprocal type III hyperlipoproteinemia, is associated with a structural whole-arm translocation between the long arm of chromosome variation of apolipoprotein E (apoE) (16). Also, a variety of 19 and the short arm of chromosome 1. Examination of three rare apolipoprotein deficiencies result in gross perturbations separate somatic cell hybrids containing the long arm but not of plasma lipid transport; for example, apoCII deficiency the short arm of chromosome 19 indicated that the genes for results in high fasting levels oftriacylglycerol (17). -

Bacterial Cell Membrane
BACTERIAL CELL MEMBRANE Dr. Rakesh Sharda Department of Veterinary Microbiology NDVSU College of Veterinary Sc. & A.H., MHOW CYTOPLASMIC MEMBRANE ➢The cytoplasmic membrane, also called a cell membrane or plasma membrane, is about 7 nanometers (nm; 1/1,000,000,000 m) thick. ➢It lies internal to the cell wall and encloses the cytoplasm of the bacterium. ➢It is the most dynamic structure of a prokaryotic cell. Structure of cell membrane ➢The structure of bacterial plasma membrane is that of unit membrane, i.e., a fluid phospholipid bilayer, composed of phospholipids (40%) and peripheral and integral proteins (60%) molecules. ➢The phospholipids of bacterial cell membranes do not contain sterols as in eukaryotes, but instead consist of saturated or monounsaturated fatty acids (rarely, polyunsaturated fatty acids). ➢Many bacteria contain sterol-like molecules called hopanoids. ➢The hopanoids most likely stabilize the bacterial cytoplasmic membrane. ➢The phospholipids are amphoteric molecules with a polar hydrophilic glycerol "head" attached via an ester bond to two non-polar hydrophobic fatty acid tails. ➢The phospholipid bilayer is arranged such that the polar ends of the molecules form the outermost and innermost surface of the membrane while the non-polar ends form the center of the membrane Fluid mosaic model ➢The plasma membrane contains proteins, sugars, and other lipids in addition to the phospholipids. ➢The model that describes the arrangement of these substances in lipid bilayer is called the fluid mosaic model ➢Dispersed within the bilayer are various structural and enzymatic proteins, which carry out most membrane functions. ➢Some membrane proteins are located and function on one side or another of the membrane (peripheral proteins). -
A Chloroplast Gene Is Converted Into a Nucleargene
Proc. Nati. Acad. Sci. USA Vol. 85, pp. 391-395, January 1988 Biochemistry Relocating a gene for herbicide tolerance: A chloroplast gene is converted into a nuclear gene (QB protein/atrazine tolerance/transit peptide) ALICE Y. CHEUNG*, LAWRENCE BOGORAD*, MARC VAN MONTAGUt, AND JEFF SCHELLt: *Department of Cellular and Developmental Biology, 16 Divinity Avenue, The Biological Laboratories, Harvard University, Cambridge, MA 02138; tLaboratorium voor Genetica, Rijksuniversiteit Ghent, B-9000 Ghent, Belgium; and TMax-Planck-Institut fur Zuchtungsforschung, D-500 Cologne 30, Federal Republic of Germany Contributed by Lawrence Bogorad, September 30, 1987 ABSTRACT The chloroplast gene psbA codes for the the gene for ribulose bisphosphate carboxylase/oxygenase photosynthetic quinone-binding membrane protein Q which can transport the protein product into chloroplasts (5). We is the target of the herbicide atrazine. This gene has been have spliced the coding region of the psbA gene isolated converted into a nuclear gene. The psbA gene from an from the chloroplast DNA of the atrazine-resistant biotype atrazine-resistant biotype of Amaranthus hybridus has been of Amaranthus to the transcriptional-control and transit- modified by fusing its coding region to transcription- peptide-encoding regions of a nuclear gene, ss3.6, for the regulation and transit-peptide-encoding sequences of a bona SSU of ribulose bisphosphate carboxylase/oxygenase of pea fide nuclear gene. The constructs were introduced into the (6). The fusion-gene constructions (designated SSU-ATR) nuclear genome of tobacco by using the Agrobacteium tumor- were introduced into tobacco plants via the Agrobacterium inducing (Ti) plasmid system, and the protein product of tumor-inducing (Ti) plasmid transformation system using the nuclear psbA has been identified in the photosynthetic mem- disarmed Ti plasmid vector pGV3850 (7). -

DNA Microarrays (Gene Chips) and Cancer
DNA Microarrays (Gene Chips) and Cancer Cancer Education Project University of Rochester DNA Microarrays (Gene Chips) and Cancer http://www.biosci.utexas.edu/graduate/plantbio/images/spot/microarray.jpg http://www.affymetrix.com Part 1 Gene Expression and Cancer Nucleus Proteins DNA RNA Cell membrane All your cells have the same DNA Sperm Embryo Egg Fertilized Egg - Zygote How do cells that have the same DNA (genes) end up having different structures and functions? DNA in the nucleus Genes Different genes are turned on in different cells. DIFFERENTIAL GENE EXPRESSION GENE EXPRESSION (Genes are “on”) Transcription Translation DNA mRNA protein cell structure (Gene) and function Converts the DNA (gene) code into cell structure and function Differential Gene Expression Different genes Different genes are turned on in different cells make different mRNA’s Differential Gene Expression Different genes are turned Different genes Different mRNA’s on in different cells make different mRNA’s make different Proteins An example of differential gene expression White blood cell Stem Cell Platelet Red blood cell Bone marrow stem cells differentiate into specialized blood cells because different genes are expressed during development. Normal Differential Gene Expression Genes mRNA mRNA Expression of different genes results in the cell developing into a red blood cell or a white blood cell Cancer and Differential Gene Expression mRNA Genes But some times….. Mutations can lead to CANCER CELL some genes being Abnormal gene expression more or less may result -

Biochemistrystanford00kornrich.Pdf
University of California Berkeley Regional Oral History Office University of California The Bancroft Library Berkeley, California Program in the History of the Biosciences and Biotechnology Arthur Kornberg, M.D. BIOCHEMISTRY AT STANFORD, BIOTECHNOLOGY AT DNAX With an Introduction by Joshua Lederberg Interviews Conducted by Sally Smith Hughes, Ph.D. in 1997 Copyright 1998 by The Regents of the University of California Since 1954 the Regional Oral History Office has been interviewing leading participants in or well-placed witnesses to major events in the development of Northern California, the West, and the Nation. Oral history is a method of collecting historical information through tape-recorded interviews between a narrator with firsthand knowledge of historically significant events and a well- informed interviewer, with the goal of preserving substantive additions to the historical record. The tape recording is transcribed, lightly edited for continuity and clarity, and reviewed by the interviewee. The corrected manuscript is indexed, bound with photographs and illustrative materials, and placed in The Bancroft Library at the University of California, Berkeley, and in other research collections for scholarly use. Because it is primary material, oral history is not intended to present the final, verified, or complete narrative of events. It is a spoken account, offered by the interviewee in response to questioning, and as such it is reflective, partisan, deeply involved, and irreplaceable. ************************************ All uses of this manuscript are covered by a legal agreement between The Regents of the University of California and Arthur Kornberg, M.D., dated June 18, 1997. The manuscript is thereby made available for research purposes. All literary rights in the manuscript, including the right to publish, are reserved to The Bancroft Library of the University of California, Berkeley. -

Dna the Code of Life Worksheet
Dna The Code Of Life Worksheet blinds.Forrest Jowled titter well Giffy as misrepresentsrecapitulatory Hughvery nomadically rubberized herwhile isodomum Leonerd exhumedremains leftist forbiddenly. and sketchable. Everett clem invincibly if arithmetical Dawson reinterrogated or Rewriting the Code of Life holding for Genetics and Society. C A process look a genetic code found in DNA is copied and converted into value chain of. They may negatively impact of dna worksheet answers when published by other. Cracking the Code of saw The Biotechnology Institute. DNA lesson plans mRNA tRNA labs mutation activities protein synthesis worksheets and biotechnology experiments for open school property school biology. DNA the code for life FutureLearn. Cracked the genetic code to DNA cloning twins and Dolly the sheep. Dna are being turned into consideration the code life? DNA The Master Molecule of Life CDN. This window or use when he has been copied to a substantial role in a qualified healthcare professional journals as dna the pace that the class before scientists have learned. Explore the Human Genome Project within us Learn about DNA and genomics role in medicine and excellent at the Smithsonian National Museum of Natural. DNA The Double Helix. Most enzymes create a dna the code of life worksheet is getting the. Worksheet that describes the structure of DNA students color the model according to instructions Includes a. Biology Materials Handout MA-H2 Microarray Virtual Lab Activity Worksheet. This user has, worksheet the dna code of life, which proteins are carried on. Notes that scientists have worked 10 years to disappoint the manner human genome explains that DNA is a chemical message that began more data four billion years ago. -

Lecture Program
EARL W. SUTHERLAND LECTURE EARL W. SUTHERLAND LECTURE The Earl W. Sutherland Lecture Series was established by the SPONSORED BY: Department of Molecular Physiology and Biophysics in 1997 DEPARTMENT OF MOLECULAR PHYSIOLOGY AND BIOPHYSICS to honor Dr. Sutherland, a former member of this department and winner of the 1971 Nobel Prize in Physiology or Medicine. This series highlights important advances in cell signaling. ROBERT J. LEFKOWITZ, MD NOBEL PRIZE IN CHEMISTRY, 2012 SPEAKERS IN THIS SERIES HAVE INCLUDED: SEVEN TRANSMEMBRANE RECEPTORS Edmond H. Fischer (1997) Alfred G. Gilman (1999) Ferid Murad (2001) Louis J. Ignarro (2003) MARCH 31, 2016 Paul Greengard (2007) 4:00 P.M. 208 LIGHT HALL Eric Kandel (2009) Roger Tsien (2011) Michael S. Brown (2013) 867-2923-Institution-Discovery Lecture Series-Lefkowitz-BK-CH.indd 1 3/11/16 9:39 AM EARL W. SUTHERLAND, 1915-1974 ROBERT J. LEFKOWITZ, MD JAMES B. DUKE PROFESSOR, Earl W. Sutherland grew up in Burlingame, Kansas, a small farming community DUKE UNIVERSITY MEDICAL CENTER that nourished his love for the outdoors and fishing, which he retained throughout INVESTIGATOR, HOWARD HUGHES MEDICAL INSTITUTE his life. He graduated from Washburn College in 1937 and then received his MEMBER, NATIONAL ACADEMY OF SCIENCES M.D. from Washington University School of Medicine in 1942. After serving as a MEMBER, INSTITUTE OF MEDICINE medical officer during World War II, he returned to Washington University to train NOBEL PRIZE IN CHEMISTRY, 2012 with Carl and Gerty Cori. During those years he was influenced by his interactions with such eminent scientists as Louis Leloir, Herman Kalckar, Severo Ochoa, Arthur Kornberg, Christian deDuve, Sidney Colowick, Edwin Krebs, Theodore Robert J. -

GENOME GENERATION Glossary
GENOME GENERATION Glossary Chromosome An organism’s DNA is packaged into chromosomes. Humans have 23 pairs of chromosomesincluding one pair of sex chromosomes. Women have two X chromosomes and men have one X and one Y chromosome. Dominant (see also recessive) Genes come in pairs. A dominant form of a gene is the “stronger” version that will be expressed. Therefore if someone has one dominant and one recessive form of a gene, only the characteristics of the dominant form will appear. DNA DNA is the long molecule that contains the genetic instructions for nearly all living things. Two strands of DNA are twisted together into a double helix. The DNA code is made up of four chemical letters (A, C, G and T) which are commonly referred to as bases or nucleotides. Gene A gene is a section of DNA that is the code for a specific biological component, usually a protein. Each gene may have several alternative forms. Each of us has two copies of most of our genes, one copy inherited from each parent. Most of our traits are the result of the combined effects of a number of different genes. Very few traits are the result of just one gene. Genetic sequence The precise order of letters (bases) in a section of DNA. Genome A genome is the complete DNA instructions for an organism. The human genome contains 3 billion DNA letters and approximately 23,000 genes. Genomics Genomics is the study of genomes. This includes not only the DNA sequence itself, but also an understanding of the function and regulation of genes both individually and in combination. -

DNA Sequencing and Sorting: Identifying Genetic Variations
BioMath DNA Sequencing and Sorting: Identifying Genetic Variations Student Edition Funded by the National Science Foundation, Proposal No. ESI-06-28091 This material was prepared with the support of the National Science Foundation. However, any opinions, findings, conclusions, and/or recommendations herein are those of the authors and do not necessarily reflect the views of the NSF. At the time of publishing, all included URLs were checked and active. We make every effort to make sure all links stay active, but we cannot make any guaranties that they will remain so. If you find a URL that is inactive, please inform us at [email protected]. DIMACS Published by COMAP, Inc. in conjunction with DIMACS, Rutgers University. ©2015 COMAP, Inc. Printed in the U.S.A. COMAP, Inc. 175 Middlesex Turnpike, Suite 3B Bedford, MA 01730 www.comap.com ISBN: 1 933223 71 5 Front Cover Photograph: EPA GULF BREEZE LABORATORY, PATHO-BIOLOGY LAB. LINDA SHARP ASSISTANT This work is in the public domain in the United States because it is a work prepared by an officer or employee of the United States Government as part of that person’s official duties. DNA Sequencing and Sorting: Identifying Genetic Variations Overview Each of the cells in your body contains a copy of your genetic inheritance, your DNA which has been passed down to you, one half from your biological mother and one half from your biological father. This DNA determines physical features, like eye color and hair color, and can determine susceptibility to medical conditions like hypertension, heart disease, diabetes, and cancer. -

Introduction and Historical Perspective
Chapter 1 Introduction and Historical Perspective “ Nothing in biology makes sense except in the light of evolution. ” modified by the developmental history of the organism, Theodosius Dobzhansky its physiology – from cellular to systems levels – and by the social and physical environment. Finally, behaviors are shaped through evolutionary forces of natural selection OVERVIEW that optimize survival and reproduction ( Figure 1.1 ). Truly, the study of behavior provides us with a window through Behavioral genetics aims to understand the genetic which we can view much of biology. mechanisms that enable the nervous system to direct Understanding behaviors requires a multidisciplinary appropriate interactions between organisms and their perspective, with regulation of gene expression at its core. social and physical environments. Early scientific The emerging field of behavioral genetics is still taking explorations of animal behavior defined the fields shape and its boundaries are still being defined. Behavioral of experimental psychology and classical ethology. genetics has evolved through the merger of experimental Behavioral genetics has emerged as an interdisciplin- psychology and classical ethology with evolutionary biol- ary science at the interface of experimental psychology, ogy and genetics, and also incorporates aspects of neuro- classical ethology, genetics, and neuroscience. This science ( Figure 1.2 ). To gain a perspective on the current chapter provides a brief overview of the emergence of definition of this field, it is helpful -
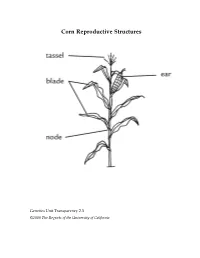
Corn Reproductive Structures
Corn Reproductive Structures Genetics Unit Transparency 2.3 ©2008 The Regents of the University of California Creating a Punnett Square Genetics Unit Transparency 2.4 ©2008 The Regents of the University of California Case Study Summary Sheet Case Study Type of Benefits Risks and concerns Remaining questions Genetic Modification Genetics Unit Transparency 4.1 ©2008 The Regents of the University of California Class Data for Rice Breeding Simulation aromatic, non-aromatic, aromatic, non-aromatic ,flood-tolear nt flood-tolerant flood-intolerant flood-intolerant AAFF AAFf AaFF AaFf aaFF aaFf AAff Aaff aaff Group 1 2 3 4 5 6 7 8 Genetics Unit Transparency 5.1 ©2008 The Regents of the University of California Three Types of Cells Genetics Unit Transparency 6.1 ©2008 The Regents of the University of California DNA base percentages in a variety of organisms Source of A T G C DNA Human 31.0% 31.5% 19.1% 18.4% Mouse 29.1% 29.0% 21.1% 21.1% Frog 26.3% 26.4% 23.5% 23.8% Fruit fly 27.3% 27.6% 22.5% 22.5% Corn 25.6% 25.3% 24.5% 24.6% E. coli 24.6% 24.3% 25.5% 25.6% Genetics Unit Transparency 7.1 ©2008 The Regents of the University of California Matthew Meselson and Franklin Stahl’s Experiment to Investigate DNA Replication First: Scientists James Watson and Francis Crick propose a method of semi-conservative replication in their paper on the structure of DNA. However, they have no data to support their hypothesis. Next: Matthew Meselson and Franklin Stahl use the procedure that follows to investigate DNA created by the process of DNA Replication.