Characterisation of Lysinibacillus Sphaericus Strains
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Alternate-Locus Aware Variant Calling in Whole Genome Sequencing Marten Jäger1,2, Max Schubach1, Tomasz Zemojtel1,Knutreinert3, Deanna M
Jäger et al. Genome Medicine (2016) 8:130 DOI 10.1186/s13073-016-0383-z RESEARCH Open Access Alternate-locus aware variant calling in whole genome sequencing Marten Jäger1,2, Max Schubach1, Tomasz Zemojtel1,KnutReinert3, Deanna M. Church4 and Peter N. Robinson1,2,3,5,6* Abstract Background: The last two human genome assemblies have extended the previous linear golden-path paradigm of the human genome to a graph-like model to better represent regions with a high degree of structural variability. The new model offers opportunities to improve the technical validity of variant calling in whole-genome sequencing (WGS). Methods: We developed an algorithm that analyzes the patterns of variant calls in the 178 structurally variable regions of the GRCh38 genome assembly, and infers whether a given sample is most likely to contain sequences from the primary assembly, an alternate locus, or their heterozygous combination at each of these 178 regions. We investigate 121 in-house WGS datasets that have been aligned to the GRCh37 and GRCh38 assemblies. Results: We show that stretches of sequences that are largely but not entirely identical between the primary assembly and an alternate locus can result in multiple variant calls against regions of the primary assembly. In WGS analysis, this results in characteristic and recognizable patterns of variant calls at positions that we term alignable scaffold-discrepant positions (ASDPs). In 121 in-house genomes, on average 51.8 ± 3.8 of the 178 regions were found to correspond best to an alternate locus rather than the primary assembly sequence, and filtering these genomes with our algorithm led to the identification of 7863 variant calls per genome that colocalized with ASDPs. -

Genome-Wide Detection of Natural Selection in African Americans Pre- and Post-Admixture
Downloaded from genome.cshlp.org on September 29, 2021 - Published by Cold Spring Harbor Laboratory Press Method Genome-wide detection of natural selection in African Americans pre- and post-admixture Wenfei Jin,1 Shuhua Xu,1,6 Haifeng Wang,2 Yongguo Yu,3 Yiping Shen,4,5 Bailin Wu,4,5 and Li Jin1,4,6 1Chinese Academy of Sciences Key Laboratory of Computational Biology, Chinese Academy of Sciences and Max Planck Society (CAS-MPG) Partner Institute for Computational Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200031, China; 2Chinese National Human Genome Center, Shanghai 201203, China; 3Shanghai Children’s Medical Center, Shanghai Jiaotong University School of Medicine, Shanghai 200127, China; 4Ministry of Education (MOE) Key Laboratory of Contemporary Anthropology, School of Life Sciences and Institutes of Biomedical Sciences, Fudan University, Shanghai 200433, China; 5Children’s Hospital Boston, Harvard Medical School, Boston, Massachusetts 02115, USA It is particularly meaningful to investigate natural selection in African Americans (AfA) due to the high mortality their African ancestry has experienced in history. In this study, we examined 491,526 autosomal single nucleotide poly- morphisms (SNPs) genotyped in 5210 individuals and conducted a genome-wide search for selection signals in 1890 AfA. Several genomic regions showing an excess of African or European ancestry, which were considered the footprints of selection since population admixture, were detected based on a commonly used approach. However, we also developed a new strategy to detect natural selection both pre- and post-admixture by reconstructing an ancestral African population (AAF) from inferred African components of ancestry in AfA and comparing it with indigenous African populations (IAF). -
Cvckc-2017-0293-0311
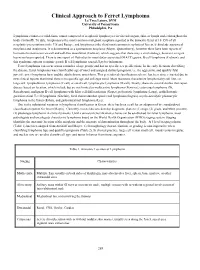
Clinical Approach to Ferret Lymphoma La’Toya Latney, DVM University of Pennsylvania Philadelphia, PA Lymphoma connotes a solid-tissue tumor composed of neoplastic lymphocytes in visceral organs, skin, or lymph nodes throughout the body (Antinoff). To date, lymphoma is the most common malignant neoplasia reported in the domestic ferret at 10-15% of all neoplastic presentations in the US and Europe, and lymphoma is the third most common neoplasia of ferrets, behind adrenocortical neoplasia and insulinoma. It is documented as a spontaneous neoplasia (Mayer, Quesenberry), however there have been reports of horizontal transmission via cell and cell-free inoculation (Erdman), which suggests that there may a viral etiology, however an agent has never been reported. There is one report of Helicobacter mustelidae-associated (MALT) gastric B-cell lymphoma (Erdman), and this syndrome appears to mimic gastric B cell lymphoma caused H.pylori in humans. Ferret lymphoma can occur across a number of age groups and has no specific sex predilections. In the early literature describing the disease, ferret lymphoma was classified by age of onset and assigned distinct prognosis, i.e. the aggressive and quickly fatal juvenile onset lymphoma form and the adult chronic onset form. This generalized classification scheme has been since retracted due to new clinical reports that reveal there is no specific age and cell-type trend. Most resources characterize lymphoma by cell line, i.e. large cell, lymphoblastic lymphoma (T cell) or small cell, lymphocytic lymphoma (B cell). Finally, there are several studies that report disease based on location, which include but are not limited to multicentric lymphoma (Ferreira), cutaneous lymphoma (Xi, Rosenbaum), malignant B-cell lymphoma with Mott cell differentiation (Gupta), polyostotic lymphoma (Long), epitheliotropic gastrointestinal T-cell lymphoma (Sinclair), focal thoracolumbar spinal cord lymphoma (Ingrao), myelo-osteolytic plasmacytic lymphoma in the femur (Eshar), and gastrointestinal lymphoma (Lee). -

MTX1 (NM 002455) Human Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC215082L3 MTX1 (NM_002455) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: MTX1 (NM_002455) Human Tagged ORF Clone Tag: Myc-DDK Symbol: MTX1 Synonyms: MTX; MTXN Vector: pLenti-C-Myc-DDK-P2A-Puro (PS100092) E. coli Selection: Chloramphenicol (34 ug/mL) Cell Selection: Puromycin ORF Nucleotide The ORF insert of this clone is exactly the same as(RC215082). Sequence: Restriction Sites: SgfI-MluI Cloning Scheme: ACCN: NM_002455 ORF Size: 1398 bp This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 MTX1 (NM_002455) Human Tagged ORF Clone – RC215082L3 OTI Disclaimer: The molecular sequence of this clone aligns with the gene accession number as a point of reference only. However, individual transcript sequences of the same gene can differ through naturally occurring variations (e.g. polymorphisms), each with its own valid existence. This clone is substantially in agreement with the reference, but a complete review of all prevailing variants is recommended prior to use. More info OTI Annotation: This clone was engineered to express the complete ORF with an expression tag. Expression varies depending on the nature of the gene. RefSeq: NM_002455.3, NP_002446.2 RefSeq Size: 1648 bp RefSeq ORF: 1401 bp Locus ID: 4580 UniProt ID: Q13505 Protein Families: Transmembrane MW: 51.5 kDa Gene Summary: Involved in transport of proteins into the mitochondrion. -

Genetic Profiling of Chromosome 1 in Breast Cancer: Mapping of Regions of Gains and Losses and Identification of Candidate Genes on 1Q
Genetic profiling of chromosome 1 in breast cancer: mapping of regions of gains and losses and identification of candidate genes on 1q. Béatrice Orsetti, Mélanie Nugoli, Nathalie Cervera, Laurence Lasorsa, Paul Chuchana, Carole Rougé, Lisa Ursule, Catherine Nguyen, Frédéric Bibeau, Carmen Rodriguez, et al. To cite this version: Béatrice Orsetti, Mélanie Nugoli, Nathalie Cervera, Laurence Lasorsa, Paul Chuchana, et al.. Genetic profiling of chromosome 1 in breast cancer: mapping of regions of gains and losses and identification of candidate genes on 1q.. British Journal of Cancer, Cancer Research UK, 2006, 95 (10), pp.1439-47. 10.1038/sj.bjc.6603433. inserm-00157275 HAL Id: inserm-00157275 https://www.hal.inserm.fr/inserm-00157275 Submitted on 25 Jun 2007 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Orsetti et al. BJC HAL author manuscript British Journal of Cancer 20/11/2006; 95(10): 1439-47 Genetic profiling of chromosome 1 in breast cancer : mapping of regions of gains and losses and identification of candidate genes HAL author manuscript inserm-00157275, version 1 on 1q Béatrice Orsetti1*, Mélanie Nugoli1*, Nathalie Cervera1, Laurence Lasorsa1, Paul Chu- chana1, Carole Rougé1, Lisa Ursule1, Catherine Nguyen2, Frédéric Bibeau3, Carmen Rodriguez1, Charles Theillet1# 1. -

Genome-Scale Crispra Screening Identifies MTX1 As a Contributor for Sorafenib Resistance in Hepatocellular Carcinoma by Augmenti
Int. J. Biol. Sci. 2021, Vol. 17 3133 Ivyspring International Publisher International Journal of Biological Sciences 2021; 17(12): 3133-3144. doi: 10.7150/ijbs.62393 Research Paper Genome-scale CRISPRa screening identifies MTX1 as a contributor for sorafenib resistance in hepatocellular carcinoma by augmenting autophagy Li Li, Shijun Yu, Qingqing Hu, Yanan Hai, Yandong Li Department of Oncology, Shanghai East Hospital, School of Medicine, Tongji University, Shanghai 200120, China Corresponding authors: Yandong Li and Yanan Hai, Department of Oncology, Shanghai East Hospital, School of Medicine, Tongji University, 150 Ji-Mo Rd., Shanghai 200120, China. Phone: 86-21-61569884; Fax: +86-21-58798999; E-mail: [email protected] and [email protected] © The author(s). This is an open access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/). See http://ivyspring.com/terms for full terms and conditions. Received: 2021.05.06; Accepted: 2021.07.09; Published: 2021.07.25 Abstract Sorafenib is the standard first-line drug for the treatment of advanced hepatocellular carcinoma (HCC), however, its therapeutic efficacy is not satisfactory due to primary or secondary resistance of HCC cells. In the present study, we identified Metaxin 1 (MTX1) as a new regulator of sorafenib resistance in HCC through genome-scale CRISPR activation (CRISPRa) screening. We found that MTX1 was frequently upregulated in HCC tissues and overexpression of MTX1 promoted HCC cell proliferation in vitro and in vivo. As well, MTX1 overexpression increased cell growth rate and decreased cell apoptosis upon sorafenib treatment. Consistently, the resistance induced by MTX1 was also observed in subcutaneous xenograft tumor model. -

Genetic Interaction Mapping and Exon-Resolution Functional Genomics with a Hybrid Cas9–Cas12a Platform
ARTICLES https://doi.org/10.1038/s41587-020-0437-z Genetic interaction mapping and exon-resolution functional genomics with a hybrid Cas9–Cas12a platform Thomas Gonatopoulos-Pournatzis 1,6 ✉ , Michael Aregger 1,6, Kevin R. Brown 1,6, Shaghayegh Farhangmehr1,2, Ulrich Braunschweig 1, Henry N. Ward 3, Kevin C. H. Ha 1, Alexander Weiss1, Maximilian Billmann 4, Tanja Durbic1, Chad L. Myers 3,4, Benjamin J. Blencowe1,2 ✉ and Jason Moffat 1,2,5 ✉ Systematic mapping of genetic interactions (GIs) and interrogation of the functions of sizable genomic segments in mammalian cells represent important goals of biomedical research. To advance these goals, we present a CRISPR (clustered regularly inter- spaced short palindromic repeats)-based screening system for combinatorial genetic manipulation that employs coexpression of CRISPR-associated nucleases 9 and 12a (Cas9 and Cas12a) and machine-learning-optimized libraries of hybrid Cas9–Cas12a guide RNAs. This system, named Cas Hybrid for Multiplexed Editing and screening Applications (CHyMErA), outperforms genetic screens using Cas9 or Cas12a editing alone. Application of CHyMErA to the ablation of mammalian paralog gene pairs reveals extensive GIs and uncovers phenotypes normally masked by functional redundancy. Application of CHyMErA in a che- mogenetic interaction screen identifies genes that impact cell growth in response to mTOR pathway inhibition. Moreover, by systematically targeting thousands of alternative splicing events, CHyMErA identifies exons underlying human cell line fit- ness. CHyMErA thus represents an effective screening approach for GI mapping and the functional analysis of sizable genomic regions, such as alternative exons. ecent breakthroughs in gene editing technologies have trans- Key to addressing these questions is the generation of tools formed mammalian cell genetics and disease research1–4. -

Chromatin Conformation Links Distal Target Genes to CKD Loci
BASIC RESEARCH www.jasn.org Chromatin Conformation Links Distal Target Genes to CKD Loci Maarten M. Brandt,1 Claartje A. Meddens,2,3 Laura Louzao-Martinez,4 Noortje A.M. van den Dungen,5,6 Nico R. Lansu,2,3,6 Edward E.S. Nieuwenhuis,2 Dirk J. Duncker,1 Marianne C. Verhaar,4 Jaap A. Joles,4 Michal Mokry,2,3,6 and Caroline Cheng1,4 1Experimental Cardiology, Department of Cardiology, Thoraxcenter Erasmus University Medical Center, Rotterdam, The Netherlands; and 2Department of Pediatrics, Wilhelmina Children’s Hospital, 3Regenerative Medicine Center Utrecht, Department of Pediatrics, 4Department of Nephrology and Hypertension, Division of Internal Medicine and Dermatology, 5Department of Cardiology, Division Heart and Lungs, and 6Epigenomics Facility, Department of Cardiology, University Medical Center Utrecht, Utrecht, The Netherlands ABSTRACT Genome-wide association studies (GWASs) have identified many genetic risk factors for CKD. However, linking common variants to genes that are causal for CKD etiology remains challenging. By adapting self-transcribing active regulatory region sequencing, we evaluated the effect of genetic variation on DNA regulatory elements (DREs). Variants in linkage with the CKD-associated single-nucleotide polymorphism rs11959928 were shown to affect DRE function, illustrating that genes regulated by DREs colocalizing with CKD-associated variation can be dysregulated and therefore, considered as CKD candidate genes. To identify target genes of these DREs, we used circular chro- mosome conformation capture (4C) sequencing on glomerular endothelial cells and renal tubular epithelial cells. Our 4C analyses revealed interactions of CKD-associated susceptibility regions with the transcriptional start sites of 304 target genes. Overlap with multiple databases confirmed that many of these target genes are involved in kidney homeostasis. -

Mouse Gba Conditional Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Gba Conditional Knockout Project (CRISPR/Cas9) Objective: To create a Gba conditional knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Gba gene (NCBI Reference Sequence: NM_008094 ; Ensembl: ENSMUSG00000028048 ) is located on Mouse chromosome 3. 12 exons are identified, with the ATG start codon in exon 3 and the TGA stop codon in exon 12 (Transcript: ENSMUST00000167998). Exon 6~8 will be selected as conditional knockout region (cKO region). Deletion of this region should result in the loss of function of the Mouse Gba gene. To engineer the targeting vector, homologous arms and cKO region will be generated by PCR using BAC clone RP23-237D17 as template. Cas9, gRNA and targeting vector will be co-injected into fertilized eggs for cKO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Mutations in this locus variably lower enzyme activity and result in accumulated glucocerebroside in reticuloendotehelial cell lysosomes and glucosylceramide in brain, liver and skin. Severe mutants die perinatally with compromised epidermal permeability. Exon 6 starts from about 25.57% of the coding region. The knockout of Exon 6~8 will result in frameshift of the gene. The size of intron 5 for 5'-loxP site insertion: 1025 bp, and the size of intron 8 for 3'-loxP site insertion: 858 bp. The size of effective cKO region: ~1422 bp. The cKO region does not have any other known gene. Page 1 of 8 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele 5' gRNA region gRNA region 3' 1 2 3 4 5 6 7 8 9 10 11 12 Targeting vector Targeted allele Constitutive KO allele (After Cre recombination) Legends Exon of mouse Gba Homology arm cKO region loxP site Page 2 of 8 https://www.alphaknockout.com Overview of the Dot Plot Window size: 10 bp Forward Reverse Complement Sequence 12 Note: The sequence of homologous arms and cKO region is aligned with itself to determine if there are tandem repeats. -

Content Based Search in Gene Expression Databases and a Meta-Analysis of Host Responses to Infection
Content Based Search in Gene Expression Databases and a Meta-analysis of Host Responses to Infection A Thesis Submitted to the Faculty of Drexel University by Francis X. Bell in partial fulfillment of the requirements for the degree of Doctor of Philosophy November 2015 c Copyright 2015 Francis X. Bell. All Rights Reserved. ii Acknowledgments I would like to acknowledge and thank my advisor, Dr. Ahmet Sacan. Without his advice, support, and patience I would not have been able to accomplish all that I have. I would also like to thank my committee members and the Biomed Faculty that have guided me. I would like to give a special thanks for the members of the bioinformatics lab, in particular the members of the Sacan lab: Rehman Qureshi, Daisy Heng Yang, April Chunyu Zhao, and Yiqian Zhou. Thank you for creating a pleasant and friendly environment in the lab. I give the members of my family my sincerest gratitude for all that they have done for me. I cannot begin to repay my parents for their sacrifices. I am eternally grateful for everything they have done. The support of my sisters and their encouragement gave me the strength to persevere to the end. iii Table of Contents LIST OF TABLES.......................................................................... vii LIST OF FIGURES ........................................................................ xiv ABSTRACT ................................................................................ xvii 1. A BRIEF INTRODUCTION TO GENE EXPRESSION............................. 1 1.1 Central Dogma of Molecular Biology........................................... 1 1.1.1 Basic Transfers .......................................................... 1 1.1.2 Uncommon Transfers ................................................... 3 1.2 Gene Expression ................................................................. 4 1.2.1 Estimating Gene Expression ............................................ 4 1.2.2 DNA Microarrays ...................................................... -

Rabbit Anti-Mitofilin Antibody-SL1824R
SunLong Biotech Co.,LTD Tel: 0086-571- 56623320 Fax:0086-571- 56623318 E-mail:[email protected] www.sunlongbiotech.com Rabbit Anti-Mitofilin antibody SL1824R Product Name: Mitofilin Chinese Name: Mitochondrion内膜蛋白抗体 Cell proliferation inducing protein 52; Cell proliferation-inducing gene 4/52 protein; HMP; IMMT; IMMT_HUMAN; Inner membrane protein mitochondrial; Inner Alias: mitochrondial membrane; MINOS2; Mitochondrial inner membrane organizing system 2; Mitochondrial inner membrane protein; Mitofilin; Motor protein; p87; p87/89; p89; pig4; PIG52; Proliferation-inducing gene 4; MICOS complex subunit MIC60. Organism Species: Rabbit Clonality: Polyclonal React Species: Human,Mouse,Rat,Dog,Cow, WB=1:500-2000ELISA=1:500-1000IHC-P=1:400-800IHC-F=1:400-800IF=1:100- 500(Paraffin sections need antigen repair) Applications: not yet tested in other applications. optimal dilutions/concentrations should be determined by the end user. Molecular weight: 68kDa Cellular localization: cytoplasmicThe cell membraneMitochondrion Form: Lyophilized or Liquid Concentration: 1mg/mlwww.sunlongbiotech.com immunogen: KLH conjugated synthetic peptide derived from human Mitofilin:241-340/758 Lsotype: IgG Purification: affinity purified by Protein A Storage Buffer: 0.01M TBS(pH7.4) with 1% BSA, 0.03% Proclin300 and 50% Glycerol. Store at -20 °C for one year. Avoid repeated freeze/thaw cycles. The lyophilized antibody is stable at room temperature for at least one month and for greater than a year Storage: when kept at -20°C. When reconstituted in sterile pH 7.4 0.01M PBS or diluent of antibody the antibody is stable for at least two weeks at 2-4 °C. PubMed: PubMed Mitochondria are the center of cellular energy production and essential metabolic Product Detail: reactions. -

Edible Insects
1.04cm spine for 208pg on 90g eco paper ISSN 0258-6150 FAO 171 FORESTRY 171 PAPER FAO FORESTRY PAPER 171 Edible insects Edible insects Future prospects for food and feed security Future prospects for food and feed security Edible insects have always been a part of human diets, but in some societies there remains a degree of disdain Edible insects: future prospects for food and feed security and disgust for their consumption. Although the majority of consumed insects are gathered in forest habitats, mass-rearing systems are being developed in many countries. Insects offer a significant opportunity to merge traditional knowledge and modern science to improve human food security worldwide. This publication describes the contribution of insects to food security and examines future prospects for raising insects at a commercial scale to improve food and feed production, diversify diets, and support livelihoods in both developing and developed countries. It shows the many traditional and potential new uses of insects for direct human consumption and the opportunities for and constraints to farming them for food and feed. It examines the body of research on issues such as insect nutrition and food safety, the use of insects as animal feed, and the processing and preservation of insects and their products. It highlights the need to develop a regulatory framework to govern the use of insects for food security. And it presents case studies and examples from around the world. Edible insects are a promising alternative to the conventional production of meat, either for direct human consumption or for indirect use as feedstock.