Guide to E. Coli Genotype and Genetic Marker Nomenclature
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Transcriptional Landscape of a Rewritten Bacterial Genome Reveals Control Elements and Genome Design Principles ✉ ✉ Mariëlle J
ARTICLE https://doi.org/10.1038/s41467-021-23362-y OPEN The transcriptional landscape of a rewritten bacterial genome reveals control elements and genome design principles ✉ ✉ Mariëlle J. F. M. van Kooten 1 , Clio A. Scheidegger1, Matthias Christen1 & Beat Christen 1 Sequence rewriting enables low-cost genome synthesis and the design of biological systems with orthogonal genetic codes. The error-free, robust rewriting of nucleotide sequences can 1234567890():,; be achieved with a complete annotation of gene regulatory elements. Here, we compare transcription in Caulobacter crescentus to transcription from plasmid-borne segments of the synthesized genome of C. ethensis 2.0. This rewritten derivative contains an extensive amount of supposedly neutral mutations, including 123’562 synonymous codon changes. The tran- scriptional landscape refines 60 promoter annotations, exposes 18 termination elements and links extensive transcription throughout the synthesized genome to the unintentional intro- duction of sigma factor binding motifs. We reveal translational regulation for 20 CDS and uncover an essential translational regulatory element for the expression of ribosomal protein RplS. The annotation of gene regulatory elements allowed us to formulate design principles that improve design schemes for synthesized DNA, en route to a bright future of iteration- free programming of biological systems. 1 Institute of Molecular Systems Biology, Department of Biology, Eidgenössische Technische Hochschule Zürich, Zürich, Switzerland. ✉ email: [email protected]; [email protected] NATURE COMMUNICATIONS | (2021) 12:3053 | https://doi.org/10.1038/s41467-021-23362-y | www.nature.com/naturecommunications 1 ARTICLE NATURE COMMUNICATIONS | https://doi.org/10.1038/s41467-021-23362-y e can program biological systems with DNA that is In previous work, we synthesized and assembled the rewritten Wbased on native nucleotide sequences. -

Development of Novel SNP Assays for Genetic Analysis of Rare Minnow (Gobiocypris Rarus) in a Successive Generation Closed Colony
diversity Article Development of Novel SNP Assays for Genetic Analysis of Rare Minnow (Gobiocypris rarus) in a Successive Generation Closed Colony Lei Cai 1,2,3, Miaomiao Hou 1,2, Chunsen Xu 1,2, Zhijun Xia 1,2 and Jianwei Wang 1,4,* 1 The Key Laboratory of Aquatic Biodiversity and Conservation of Chinese Academy of Sciences, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430070, China; [email protected] (L.C.); [email protected] (M.H.); [email protected] (C.X.); [email protected] (Z.X.) 2 University of Chinese Academy of Sciences, Beijing 100049, China 3 Guangdong Provincial Key Laboratory of Laboratory Animals, Guangdong Laboratory Animals Monitoring Institute, Guangzhou 510663, China 4 National Aquatic Biological Resource Center, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430070, China * Correspondence: [email protected] Received: 10 November 2020; Accepted: 11 December 2020; Published: 18 December 2020 Abstract: The complex genetic architecture of closed colonies during successive passages poses a significant challenge in the understanding of the genetic background. Research on the dynamic changes in genetic structure for the establishment of a new closed colony is limited. In this study, we developed 51 single nucleotide polymorphism (SNP) markers for the rare minnow (Gobiocypris rarus) and conducted genetic diversity and structure analyses in five successive generations of a closed colony using 20 SNPs. The range of mean Ho and He in five generations was 0.4547–0.4983 and 0.4445–0.4644, respectively. No significant differences were observed in the Ne, Ho, and He (p > 0.05) between the five closed colony generations, indicating well-maintained heterozygosity. -

Evidence That Tolc Is Required for Functioning of the Mar/Acrab Efffux
JOURNAL OF BACTERIOLOGY, Oct. 1996, p. 5803–5805 Vol. 178, No. 19 0021-9193/96/$04.0010 Copyright q 1996, American Society for Microbiology Evidence that TolC Is Required for Functioning of the Mar/AcrAB Efflux Pump of Escherichia coli JOE A. FRALICK* Department of Microbiology and Immunology, Texas Tech University Health Sciences Center, Lubbock, Texas 79403 Received 10 June 1996/Accepted 16 July 1996 A study examining the influence of TolC on AcrA, AcrR, and MarR1 mutants indicates that functional TolC is required for the operation of the AcrAB efflux system and for the expression of the Mar phenotype. That the effect of TolC on the AcrAB pump is not regulatory in nature is shown by studies measuring the influence of a tolC::Tn10 insertion mutation on the expression of an acrA::lacZ reporter fusion. These results are compatible with the hypothesis that TolC is a component of the AcrAB efflux complex. Enteric bacteria have evolved to survive in an environment the heptose I phosphate of TolC LPS may be blocked by a rich with pernicious agents, such as bile salts, detergents, and phosphorylethanolamine group (32), it has been suggested that fatty acids. Protection against these inhibitors is due, in part, to the hypersusceptibility seen in TolC mutants is the result of the the selective permeability barrier of an outer membrane, which inability of TolC LPS to form a highly ordered LPS monolayer serves as the first line of defense against the rapid intrusion of (32). There are, however, significant differences between the lipophilic compounds (2, 11, 24, 25, 27). -

Genetic Linkage Analysis
BASIC SCIENCE SEMINARS IN NEUROLOGY SECTION EDITOR: HASSAN M. FATHALLAH-SHAYKH, MD Genetic Linkage Analysis Stefan M. Pulst, MD enetic linkage analysis is a powerful tool to detect the chromosomal location of dis- ease genes. It is based on the observation that genes that reside physically close on a chromosome remain linked during meiosis. For most neurologic diseases for which the underlying biochemical defect was not known, the identification of the chromo- Gsomal location of the disease gene was the first step in its eventual isolation. By now, genes that have been isolated in this way include examples from all types of neurologic diseases, from neu- rodegenerative diseases such as Alzheimer, Parkinson, or ataxias, to diseases of ion channels lead- ing to periodic paralysis or hemiplegic migraine, to tumor syndromes such as neurofibromatosis types 1 and 2. Arch Neurol. 1999;56:667-672 With the advent of new genetic markers tin gene, diagnosis using flanking mark- and automated genotyping, genetic map- ers requires the analysis of several family ping can be conducted extremely rap- members. idly. Genetic linkage maps have been gen- erated for the human genome and for LINKAGE OF GENES model organisms and have provided the basis for the construction of physical maps When Mendel observed an “independent that permit the rapid mapping of disease assortment of traits” (Mendel’s second traits. law), he was fortunate to have chosen traits As soon as a chromosomal location that were not localized close to one an- for a disease phenotype has been estab- other on the same chromosome.1 Subse- lished, genetic linkage analysis helps quent studies revealed that many genes determine whether the disease pheno- were indeed linked, ie, that traits did not type is only caused by mutation in a assort or segregate independently, but that single gene or mutations in other genes traits encoded by these linked genes were can give rise to an identical or similar inherited together. -

Descriptors for Genetic Markers Technologies
• Descriptors for genetic markers technologies Version 1.0, February 2004 Carmen De Vicente, Thomas Metz and Adriana Alercia <www.futureharvest.org> IPGRI is a Future Harvest Centre supported by the Consultative Group on International Agricultural Research (CGIAR) ii DESCRIPTORS FOR GENETIC MARKERS TECHNOLOGIES The International Plant Genetic Resources Institute (IPGRI) is an independent international scientific organization that seeks to advance the conservation and use of plant genetic diversity for the well-being of present and future generations. It is one of 16 Future Harvest Centres supported by the Consultative Group on International Agricultural Research (CGIAR), an association of public and private members who support efforts to mobilize cutting-edge science to reduce hunger and poverty, improve human nutrition and health, and protect the environment. IPGRI has its headquarters in Maccarese, near Rome, Italy, with offices in more than 20 other countries worldwide. The Institute operates through three programmes: (1) the Plant Genetic Resources Programme, (2) the CGIAR Genetic Resources Support Programme and (3) the International Network for the Improvement of Banana and Plantain (INIBAP). The international status of IPGRI is conferred under an Establishment Agreement which, by January 2003, had been signed by the Governments of Algeria, Australia, Belgium, Benin, Bolivia, Brazil, Burkina Faso, Cameroon, Chile, China, Congo, Costa Rica, Côte d’Ivoire, Cyprus, Czech Republic, Denmark, Ecuador, Egypt, Greece, Guinea, Hungary, India, Indonesia, Iran, Israel, Italy, Jordan, Kenya, Malaysia, Mauritania, Morocco, Norway, Pakistan, Panama, Peru, Poland, Portugal, Romania, Russia, Senegal, Slovakia, Sudan, Switzerland, Syria, Tunisia, Turkey, Uganda and Ukraine. Financial support for IPGRI’s research is provided by more than 150 donors, including governments, private foundations and international organizations. -

Horizontal Gene Flow Into Geobacillus Is Constrained by the Chromosomal Organization of Growth and Sporulation
bioRxiv preprint doi: https://doi.org/10.1101/381442; this version posted August 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Horizontal gene flow into Geobacillus is constrained by the chromosomal organization of growth and sporulation Alexander Esin1,2, Tom Ellis3,4, Tobias Warnecke1,2* 1Molecular Systems Group, Medical Research Council London Institute of Medical Sciences, London, United Kingdom 2Institute of Clinical Sciences, Faculty of Medicine, Imperial College London, London, United Kingdom 3Imperial College Centre for Synthetic Biology, Imperial College London, London, United Kingdom 4Department of Bioengineering, Imperial College London, London, United Kingdom *corresponding author ([email protected]) 1 bioRxiv preprint doi: https://doi.org/10.1101/381442; this version posted August 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Abstract Horizontal gene transfer (HGT) in bacteria occurs in the context of adaptive genome architecture. As a consequence, some chromosomal neighbourhoods are likely more permissive to HGT than others. Here, we investigate the chromosomal topology of horizontal gene flow into a clade of Bacillaceae that includes Geobacillus spp. Reconstructing HGT patterns using a phylogenetic approach coupled to model-based reconciliation, we discover three large contiguous chromosomal zones of HGT enrichment. -

Section 4. Guidance Document on Horizontal Gene Transfer Between Bacteria
306 - PART 2. DOCUMENTS ON MICRO-ORGANISMS Section 4. Guidance document on horizontal gene transfer between bacteria 1. Introduction Horizontal gene transfer (HGT) 1 refers to the stable transfer of genetic material from one organism to another without reproduction. The significance of horizontal gene transfer was first recognised when evidence was found for ‘infectious heredity’ of multiple antibiotic resistance to pathogens (Watanabe, 1963). The assumed importance of HGT has changed several times (Doolittle et al., 2003) but there is general agreement now that HGT is a major, if not the dominant, force in bacterial evolution. Massive gene exchanges in completely sequenced genomes were discovered by deviant composition, anomalous phylogenetic distribution, great similarity of genes from distantly related species, and incongruent phylogenetic trees (Ochman et al., 2000; Koonin et al., 2001; Jain et al., 2002; Doolittle et al., 2003; Kurland et al., 2003; Philippe and Douady, 2003). There is also much evidence now for HGT by mobile genetic elements (MGEs) being an ongoing process that plays a primary role in the ecological adaptation of prokaryotes. Well documented is the example of the dissemination of antibiotic resistance genes by HGT that allowed bacterial populations to rapidly adapt to a strong selective pressure by agronomically and medically used antibiotics (Tschäpe, 1994; Witte, 1998; Mazel and Davies, 1999). MGEs shape bacterial genomes, promote intra-species variability and distribute genes between distantly related bacterial genera. Horizontal gene transfer (HGT) between bacteria is driven by three major processes: transformation (the uptake of free DNA), transduction (gene transfer mediated by bacteriophages) and conjugation (gene transfer by means of plasmids or conjugative and integrated elements). -

Single Nucleotide Polymorphism (SNP) Discovery in Mammals
Molecular Ecology (2004) 13, 1423–1431 doi: 10.1111/j.1365-294X.2004.02159.x SingleBlackwell Publishing, Ltd. nucleotide polymorphism (SNP) discovery in mammals: a targeted-gene approach NICOLA AITKEN,* STEVEN SMITH,† CARSTEN SCHWARZ‡ and PHILLIP A. MORIN§ Laboratory for Conservation Genetics, Max Planck Institute for Evolutionary Anthropology, Inselstrasse 22, D-04103, Leipzig, Germany Abstract Single nucleotide polymorphisms (SNPs) have rarely been exploited in nonhuman and nonmodel organism genetic studies. This is due partly to difficulties in finding SNPs in species where little DNA sequence data exist, as well as to a lack of robust and inexpensive genotyping methods. We have explored one SNP discovery method for molecular ecology, evolution, and conservation studies to evaluate the method and its limitations for population genetics in mammals. We made use of ‘CATS’ (or ‘EPIC’) primers to screen for novel SNPs in mammals. Most of these primer sets were designed from primates and/or rodents, for amplifying intron regions from conserved genes. We have screened 202 loci in 16 repres- entatives of the major mammalian clades. Polymerase chain reaction (PCR) success correlated with phylogenetic distance from the human and mouse sequences used to design most primers; for example, specific PCR products from primates and the mouse amplified the most con- sistently and the marsupial and armadillo amplifications were least successful. Approxi- mately 24% (opossum) to 65% (chimpanzee) of primers produced usable PCR product(s) in the mammals tested. Products produced generally high but variable levels of readable sequence and similarity to the expected genes. In a preliminary screen of chimpanzee DNA, 12 SNPs were identified from six (of 11) sequenced regions, yielding a SNP on average every 400 base pairs (bp). -

Roper, Hemmons
ORIGINS OF TRANSLOCATIONS IN ASPERGZLLUS NZDULANSl ETTA KAFER Department of Genetics, McGill University, Montreal, Cam& T was realized early in the course of the genetic analysis of the ascomycete ‘Aspergillus nidukns that several of the strains with X-ray induced mutants contain chromosomal aberrations (e.g. two cases mentioned by PONTECORVO, ROPER,HEMMONS, MACDONALD and BUFTON1953). At that time all evidence came from meiotic analysis. In crosses heterozygous for chromosomal aberra- tions, crossing over appears to be reduced since certain unbalanced crossover types show low viability. In fungi, this results in an unusually high frequency of aborted and abnormal ascospores, corresponding to defective pollen in higher plants that show “semisterility” as a result of chromosome aberrations. Ascopore patterns can be used to detect visually the presence of aberrations in Neurospora, either in intact asci (MCCLINTOCK1945), or in random spores (PERKINS, GLASSEYand BLOOM1962). The nonlinear ascus of Aspergillus is too small to make use of this method routinely even though similar patterns have been ob- served in asci from a cross which is now known to have been heterozygous for at least three aberrations (ELLIOTT1960). On the other hand, the discovery of mitotic recombination in Aspergillus (ROPER1952; PONTECORVOand ROPER 1953) has provided new genetic methods not only for the mapping of markers but also for the detection of aberrations, especially translocations (PONTECORVO and =FER 1958; KAFER 1958). Even though most X-ray mutants had been excluded from the general Glasgow stocks, several aberrations have been encountered in the course of mitotic mapping of new markers. The first of these resulted in mitotic linkage between markers of linkage groups I and VII. -

Basic Genetic Terms for Teachers
Student Name: Date: Class Period: Page | 1 Basic Genetic Terms Use the available reference resources to complete the table below. After finding out the definition of each word, rewrite the definition using your own words (middle column), and provide an example of how you may use the word (right column). Genetic Terms Definition in your own words An example Allele Different forms of a gene, which produce Different alleles produce different hair colors—brown, variations in a genetically inherited trait. blond, red, black, etc. Genes Genes are parts of DNA and carry hereditary Genes contain blue‐print for each individual for her or information passed from parents to children. his specific traits. Dominant version (allele) of a gene shows its Dominant When a child inherits dominant brown‐hair gene form specific trait even if only one parent passed (allele) from dad, the child will have brown hair. the gene to the child. When a child inherits recessive blue‐eye gene form Recessive Recessive gene shows its specific trait when (allele) from both mom and dad, the child will have blue both parents pass the gene to the child. eyes. Homozygous Two of the same form of a gene—one from Inheriting the same blue eye gene form from both mom and the other from dad. parents result in a homozygous gene. Heterozygous Two different forms of a gene—one from Inheriting different eye color gene forms from mom mom and the other from dad are different. and dad result in a heterozygous gene. Genotype Internal heredity information that contain Blue eye and brown eye have different genotypes—one genetic code. -

Molecular Biology and Applied Genetics
MOLECULAR BIOLOGY AND APPLIED GENETICS FOR Medical Laboratory Technology Students Upgraded Lecture Note Series Mohammed Awole Adem Jimma University MOLECULAR BIOLOGY AND APPLIED GENETICS For Medical Laboratory Technician Students Lecture Note Series Mohammed Awole Adem Upgraded - 2006 In collaboration with The Carter Center (EPHTI) and The Federal Democratic Republic of Ethiopia Ministry of Education and Ministry of Health Jimma University PREFACE The problem faced today in the learning and teaching of Applied Genetics and Molecular Biology for laboratory technologists in universities, colleges andhealth institutions primarily from the unavailability of textbooks that focus on the needs of Ethiopian students. This lecture note has been prepared with the primary aim of alleviating the problems encountered in the teaching of Medical Applied Genetics and Molecular Biology course and in minimizing discrepancies prevailing among the different teaching and training health institutions. It can also be used in teaching any introductory course on medical Applied Genetics and Molecular Biology and as a reference material. This lecture note is specifically designed for medical laboratory technologists, and includes only those areas of molecular cell biology and Applied Genetics relevant to degree-level understanding of modern laboratory technology. Since genetics is prerequisite course to molecular biology, the lecture note starts with Genetics i followed by Molecular Biology. It provides students with molecular background to enable them to understand and critically analyze recent advances in laboratory sciences. Finally, it contains a glossary, which summarizes important terminologies used in the text. Each chapter begins by specific learning objectives and at the end of each chapter review questions are also included. -
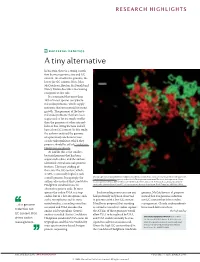
Bacterial Genetics a Tiny Alternative
RESEARCH HIGHLIGHTS BACTERIAL GENETICS A tiny alternative In bacteria, there is a strong correla- tion between genome size and GC content: the smaller the genome, the lower the GC content. Now, John McCutcheon, Bradon McDonald and Nancy Moran describe a fascinating exception to this rule. It is estimated that more than 10% of insect species carry bacte- rial endosymbionts, which supply nutrients that are essential for insect growth. The genomes of the bacte- rial endosymbionts that have been sequenced so far are much smaller than the genomes of other intracel- lular or free-living bacteria and all have a low GC content. In this study, the authors analysed the genome of a previously uncharacterized cicada endosymbiont, which they propose should be called Candidatus Hodgkinia cicadicola. At 144 kb, this is the smallest bacterial genome that has been sequenced to date, and the authors identified several unusual genomic features. The most striking of these was the GC content, which, at 58%, is unusually high for such Micrograph showing Candidatus Hodgkinia cicadicola (red) in close association with another endosymbiont, a small genome. Intriguingly, the Candidatus Sulcia muelleri (green) in the cicada Diceroprocta semicincta. The scale bar represents 10 µm. authors also noticed that Candidatus Image reproduced from McCutcheon, J. P., McDonald, B. R. & Moran, N. P. Origin of an alternative genetic Hodgkinia cicadicola uses an code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 5, e1000565 (2009). alternative genetic code. In most species the codon UGA is a stop Such recoding events are rare and genome, McCutcheon et al.