A Cladistic Analysis of Graecopithecus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Place of the Neanderthals in Hominin Phylogeny ⇑ Suzanna White, John A.J
Journal of Anthropological Archaeology 35 (2014) 32–50 Contents lists available at ScienceDirect Journal of Anthropological Archaeology journal homepage: www.elsevier.com/locate/jaa The place of the Neanderthals in hominin phylogeny ⇑ Suzanna White, John A.J. Gowlett, Matt Grove School of Archaeology, Classics and Egyptology, University of Liverpool, William Hartley Building, Brownlow Street, Liverpool L69 3GS, UK article info abstract Article history: Debate over the taxonomic status of the Neanderthals has been incessant since the initial discovery of the Received 8 May 2013 type specimens, with some arguing they should be included within our species (i.e. Homo sapiens nean- Revision received 21 March 2014 derthalensis) and others believing them to be different enough to constitute their own species (Homo Available online 13 May 2014 neanderthalensis). This synthesis addresses the process of speciation as well as incorporating information on the differences between species and subspecies, and the criteria used for discriminating between the Keywords: two. It also analyses the evidence for Neanderthal–AMH hybrids, and their relevance to the species Neanderthals debate, before discussing morphological and genetic evidence relevant to the Neanderthal taxonomic Taxonomy debate. The main conclusion is that Neanderthals fulfil all major requirements for species status. The Species status extent of interbreeding between the two populations is still highly debated, and is irrelevant to the issue at hand, as the Biological Species Concept allows -
Hands-On Human Evolution: a Laboratory Based Approach
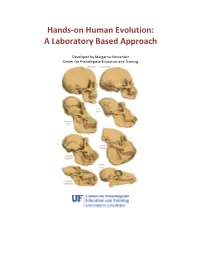
Hands-on Human Evolution: A Laboratory Based Approach Developed by Margarita Hernandez Center for Precollegiate Education and Training Author: Margarita Hernandez Curriculum Team: Julie Bokor, Sven Engling A huge thank you to….. Contents: 4. Author’s note 5. Introduction 6. Tips about the curriculum 8. Lesson Summaries 9. Lesson Sequencing Guide 10. Vocabulary 11. Next Generation Sunshine State Standards- Science 12. Background information 13. Lessons 122. Resources 123. Content Assessment 129. Content Area Expert Evaluation 131. Teacher Feedback Form 134. Student Feedback Form Lesson 1: Hominid Evolution Lab 19. Lesson 1 . Student Lab Pages . Student Lab Key . Human Evolution Phylogeny . Lab Station Numbers . Skeletal Pictures Lesson 2: Chromosomal Comparison Lab 48. Lesson 2 . Student Activity Pages . Student Lab Key Lesson 3: Naledi Jigsaw 77. Lesson 3 Author’s note Introduction Page The validity and importance of the theory of biological evolution runs strong throughout the topic of biology. Evolution serves as a foundation to many biological concepts by tying together the different tenants of biology, like ecology, anatomy, genetics, zoology, and taxonomy. It is for this reason that evolution plays a prominent role in the state and national standards and deserves thorough coverage in a classroom. A prime example of evolution can be seen in our own ancestral history, and this unit provides students with an excellent opportunity to consider the multiple lines of evidence that support hominid evolution. By allowing students the chance to uncover the supporting evidence for evolution themselves, they discover the ways the theory of evolution is supported by multiple sources. It is our hope that the opportunity to handle our ancestors’ bone casts and examine real molecular data, in an inquiry based environment, will pique the interest of students, ultimately leading them to conclude that the evidence they have gathered thoroughly supports the theory of evolution. -

Download Full Article in PDF Format
Dryopithecins, Darwin, de Bonis, and the European origin of the African apes and human clade David R. BEGUN University of Toronto, Department of Anthropology, 19 Russell Street, Toronto, ON, M5S 2S2 (Canada) [email protected] Begun R. D. 2009. — Dryopithecins, Darwin, de Bonis, and the European origin of the African apes and human clade. Geodiversitas 31 (4) : 789-816. ABSTRACT Darwin famously opined that the most likely place of origin of the common ancestor of African apes and humans is Africa, given the distribution of its liv- ing descendents. But it is infrequently recalled that immediately afterwards, Darwin, in his typically thorough and cautious style, noted that a fossil ape from Europe, Dryopithecus, may instead represent the ancestors of African apes, which dispersed into Africa from Europe. Louis de Bonis and his collaborators were the fi rst researchers in the modern era to echo Darwin’s suggestion about apes from Europe. Resulting from their spectacular discoveries in Greece over several decades, de Bonis and colleagues have shown convincingly that African KEY WORDS ape and human clade members (hominines) lived in Europe at least 9.5 million Mammalia, years ago. Here I review the fossil record of hominoids in Europe as it relates to Primates, Dryopithecus, the origins of the hominines. While I diff er in some details with Louis, we are Hispanopithecus, in complete agreement on the importance of Europe in determining the fate Rudapithecus, of the African ape and human clade. Th ere is no doubt that Louis de Bonis is Ouranopithecus, hominine origins, a pioneer in advancing our understanding of this fascinating time in our evo- new subtribe. -

Human Evolution: a Paleoanthropological Perspective - F.H
PHYSICAL (BIOLOGICAL) ANTHROPOLOGY - Human Evolution: A Paleoanthropological Perspective - F.H. Smith HUMAN EVOLUTION: A PALEOANTHROPOLOGICAL PERSPECTIVE F.H. Smith Department of Anthropology, Loyola University Chicago, USA Keywords: Human evolution, Miocene apes, Sahelanthropus, australopithecines, Australopithecus afarensis, cladogenesis, robust australopithecines, early Homo, Homo erectus, Homo heidelbergensis, Australopithecus africanus/Australopithecus garhi, mitochondrial DNA, homology, Neandertals, modern human origins, African Transitional Group. Contents 1. Introduction 2. Reconstructing Biological History: The Relationship of Humans and Apes 3. The Human Fossil Record: Basal Hominins 4. The Earliest Definite Hominins: The Australopithecines 5. Early Australopithecines as Primitive Humans 6. The Australopithecine Radiation 7. Origin and Evolution of the Genus Homo 8. Explaining Early Hominin Evolution: Controversy and the Documentation- Explanation Controversy 9. Early Homo erectus in East Africa and the Initial Radiation of Homo 10. After Homo erectus: The Middle Range of the Evolution of the Genus Homo 11. Neandertals and Late Archaics from Africa and Asia: The Hominin World before Modernity 12. The Origin of Modern Humans 13. Closing Perspective Glossary Bibliography Biographical Sketch Summary UNESCO – EOLSS The basic course of human biological history is well represented by the existing fossil record, although there is considerable debate on the details of that history. This review details both what is firmly understood (first echelon issues) and what is contentious concerning humanSAMPLE evolution. Most of the coCHAPTERSntention actually concerns the details (second echelon issues) of human evolution rather than the fundamental issues. For example, both anatomical and molecular evidence on living (extant) hominoids (apes and humans) suggests the close relationship of African great apes and humans (hominins). That relationship is demonstrated by the existing hominoid fossil record, including that of early hominins. -

The Threads of Evolutionary, Behavioural and Conservation Research
Taxonomic Tapestries The Threads of Evolutionary, Behavioural and Conservation Research Taxonomic Tapestries The Threads of Evolutionary, Behavioural and Conservation Research Edited by Alison M Behie and Marc F Oxenham Chapters written in honour of Professor Colin P Groves Published by ANU Press The Australian National University Acton ACT 2601, Australia Email: [email protected] This title is also available online at http://press.anu.edu.au National Library of Australia Cataloguing-in-Publication entry Title: Taxonomic tapestries : the threads of evolutionary, behavioural and conservation research / Alison M Behie and Marc F Oxenham, editors. ISBN: 9781925022360 (paperback) 9781925022377 (ebook) Subjects: Biology--Classification. Biology--Philosophy. Human ecology--Research. Coexistence of species--Research. Evolution (Biology)--Research. Taxonomists. Other Creators/Contributors: Behie, Alison M., editor. Oxenham, Marc F., editor. Dewey Number: 578.012 All rights reserved. No part of this publication may be reproduced, stored in a retrieval system or transmitted in any form or by any means, electronic, mechanical, photocopying or otherwise, without the prior permission of the publisher. Cover design and layout by ANU Press Cover photograph courtesy of Hajarimanitra Rambeloarivony Printed by Griffin Press This edition © 2015 ANU Press Contents List of Contributors . .vii List of Figures and Tables . ix PART I 1. The Groves effect: 50 years of influence on behaviour, evolution and conservation research . 3 Alison M Behie and Marc F Oxenham PART II 2 . Characterisation of the endemic Sulawesi Lenomys meyeri (Muridae, Murinae) and the description of a new species of Lenomys . 13 Guy G Musser 3 . Gibbons and hominoid ancestry . 51 Peter Andrews and Richard J Johnson 4 . -

Paranthropus Boisei: Fifty Years of Evidence and Analysis Bernard A
Marshall University Marshall Digital Scholar Biological Sciences Faculty Research Biological Sciences Fall 11-28-2007 Paranthropus boisei: Fifty Years of Evidence and Analysis Bernard A. Wood George Washington University Paul J. Constantino Biological Sciences, [email protected] Follow this and additional works at: http://mds.marshall.edu/bio_sciences_faculty Part of the Biological and Physical Anthropology Commons Recommended Citation Wood B and Constantino P. Paranthropus boisei: Fifty years of evidence and analysis. Yearbook of Physical Anthropology 50:106-132. This Article is brought to you for free and open access by the Biological Sciences at Marshall Digital Scholar. It has been accepted for inclusion in Biological Sciences Faculty Research by an authorized administrator of Marshall Digital Scholar. For more information, please contact [email protected], [email protected]. YEARBOOK OF PHYSICAL ANTHROPOLOGY 50:106–132 (2007) Paranthropus boisei: Fifty Years of Evidence and Analysis Bernard Wood* and Paul Constantino Center for the Advanced Study of Hominid Paleobiology, George Washington University, Washington, DC 20052 KEY WORDS Paranthropus; boisei; aethiopicus; human evolution; Africa ABSTRACT Paranthropus boisei is a hominin taxon ers can trace the evolution of metric and nonmetric var- with a distinctive cranial and dental morphology. Its iables across hundreds of thousands of years. This pa- hypodigm has been recovered from sites with good per is a detailed1 review of half a century’s worth of fos- stratigraphic and chronological control, and for some sil evidence and analysis of P. boi se i and traces how morphological regions, such as the mandible and the both its evolutionary history and our understanding of mandibular dentition, the samples are not only rela- its evolutionary history have evolved during the past tively well dated, but they are, by paleontological 50 years. -

Unravelling the Positional Behaviour of Fossil Hominoids: Morphofunctional and Structural Analysis of the Primate Hindlimb
ADVERTIMENT. Lʼaccés als continguts dʼaquesta tesi queda condicionat a lʼacceptació de les condicions dʼús establertes per la següent llicència Creative Commons: http://cat.creativecommons.org/?page_id=184 ADVERTENCIA. El acceso a los contenidos de esta tesis queda condicionado a la aceptación de las condiciones de uso establecidas por la siguiente licencia Creative Commons: http://es.creativecommons.org/blog/licencias/ WARNING. The access to the contents of this doctoral thesis it is limited to the acceptance of the use conditions set by the following Creative Commons license: https://creativecommons.org/licenses/?lang=en Doctorado en Biodiversitat Facultad de Ciènces Tesis doctoral Unravelling the positional behaviour of fossil hominoids: Morphofunctional and structural analysis of the primate hindlimb Marta Pina Miguel 2016 Memoria presentada por Marta Pina Miguel para optar al grado de Doctor por la Universitat Autònoma de Barcelona, programa de doctorado en Biodiversitat del Departamento de Biologia Animal, de Biologia Vegetal i d’Ecologia (Facultad de Ciències). Este trabajo ha sido dirigido por el Dr. Salvador Moyà Solà (Institut Català de Paleontologia Miquel Crusafont) y el Dr. Sergio Almécija Martínez (The George Washington Univertisy). Director Co-director Dr. Salvador Moyà Solà Dr. Sergio Almécija Martínez A mis padres y hermana. Y a todas aquelas personas que un día decidieron perseguir un sueño Contents Acknowledgments [in Spanish] 13 Abstract 19 Resumen 21 Section I. Introduction 23 Hominoid positional behaviour The great apes of the Vallès-Penedès Basin: State-of-the-art Section II. Objectives 55 Section III. Material and Methods 59 Hindlimb fossil remains of the Vallès-Penedès hominoids Comparative sample Area of study: The Vallès-Penedès Basin Methodology: Generalities and principles Section IV. -

Chapter 1 - Introduction
EURASIAN MIDDLE AND LATE MIOCENE HOMINOID PALEOBIOGEOGRAPHY AND THE GEOGRAPHIC ORIGINS OF THE HOMININAE by Mariam C. Nargolwalla A thesis submitted in conformity with the requirements for the degree of Doctor of Philosophy Graduate Department of Anthropology University of Toronto © Copyright by M. Nargolwalla (2009) Eurasian Middle and Late Miocene Hominoid Paleobiogeography and the Geographic Origins of the Homininae Mariam C. Nargolwalla Doctor of Philosophy Department of Anthropology University of Toronto 2009 Abstract The origin and diversification of great apes and humans is among the most researched and debated series of events in the evolutionary history of the Primates. A fundamental part of understanding these events involves reconstructing paleoenvironmental and paleogeographic patterns in the Eurasian Miocene; a time period and geographic expanse rich in evidence of lineage origins and dispersals of numerous mammalian lineages, including apes. Traditionally, the geographic origin of the African ape and human lineage is considered to have occurred in Africa, however, an alternative hypothesis favouring a Eurasian origin has been proposed. This hypothesis suggests that that after an initial dispersal from Africa to Eurasia at ~17Ma and subsequent radiation from Spain to China, fossil apes disperse back to Africa at least once and found the African ape and human lineage in the late Miocene. The purpose of this study is to test the Eurasian origin hypothesis through the analysis of spatial and temporal patterns of distribution, in situ evolution, interprovincial and intercontinental dispersals of Eurasian terrestrial mammals in response to environmental factors. Using the NOW and Paleobiology databases, together with data collected through survey and excavation of middle and late Miocene vertebrate localities in Hungary and Romania, taphonomic bias and sampling completeness of Eurasian faunas are assessed. -

Origin of Language
Origin of language The origin of language in the human species has been the topic of scholarly discussions for several centuries. There is no consensus on the origin or age of human language. The topic is difficult to study because of the lack of direct evidence. Consequently, scholars wishing to study the origins of language must draw inferences from other kinds of evidence such as the fossil record, archaeological evidence, contemporary language diversity, studies of language acquisition, and comparisons between human language and systems of communication existing among other animals (particularly other primates). Many argue that the origins of language probably relate closely to the origins of modern human behavior, but there is little agreement about the implications and directionality of this connection. This shortage of empirical evidence has led many scholars to regard the entire topic as unsuitable for serious study. In 1866, the Linguistic Society of Paris banned any existing or future debates on the subject, a prohibition which remained influential across much of the western world until late in the twentieth century.[1] Today, there are various hypotheses about how, why, when, and where language might have emerged.[2] Despite this, there is scarcely more agreement today than a hundred years ago, when Charles Darwin's theory of evolution by natural selection provoked a rash of armchair speculation on the topic.[3] Since the early 1990s, however, a number of linguists, archaeologists, psychologists, anthropologists, and others -

Environmental Hypotheses of Hominin Evolution
YEARBOOK OF PHYSICAL ANTHROPOLOGY 41:93–136 (1998) Environmental Hypotheses of Hominin Evolution RICHARD POTTS Human Origins Program, National Museum of Natural History, Smithsonian Institution, Washington, DC 20560-0112 KEY WORDS climate; habitat; adaptation; variability selection; fossil humans; stone tools ABSTRACT The study of human evolution has long sought to explain major adaptations and trends that led to the origin of Homo sapiens. Environmental scenarios have played a pivotal role in this endeavor. They represent statements or, more commonly, assumptions concerning the adap- tive context in which key hominin traits emerged. In many cases, however, these scenarios are based on very little if any data about the past settings in which early hominins lived. Several environmental hypotheses of human evolution are presented in this paper. Explicit test expectations are laid out, and a preliminary assessment of the hypotheses is made by examining the environmental records of Olduvai, Turkana, Olorgesailie, Zhoukoudian, Combe Grenal, and other hominin localities. Habitat-specific hypotheses have pre- vailed in almost all previous accounts of human adaptive history. The rise of African dry savanna is often cited as the critical event behind the develop- ment of terrestrial bipedality, stone toolmaking, and encephalized brains, among other traits. This savanna hypothesis has been countered recently by the woodland/forest hypothesis, which claims that Pliocene hominins had evolved in and were primarily attracted to closed habitats. The ideas that human evolution was fostered by cold habitats in higher latitudes or by seasonal variations in tropical and temperate zones also have their propo- nents. An alternative view, the variability selection hypothesis, states that large disparities in environmental conditions were responsible for important episodes of adaptive evolution. -

71St Annual Meeting Society of Vertebrate Paleontology Paris Las Vegas Las Vegas, Nevada, USA November 2 – 5, 2011 SESSION CONCURRENT SESSION CONCURRENT
ISSN 1937-2809 online Journal of Supplement to the November 2011 Vertebrate Paleontology Vertebrate Society of Vertebrate Paleontology Society of Vertebrate 71st Annual Meeting Paleontology Society of Vertebrate Las Vegas Paris Nevada, USA Las Vegas, November 2 – 5, 2011 Program and Abstracts Society of Vertebrate Paleontology 71st Annual Meeting Program and Abstracts COMMITTEE MEETING ROOM POSTER SESSION/ CONCURRENT CONCURRENT SESSION EXHIBITS SESSION COMMITTEE MEETING ROOMS AUCTION EVENT REGISTRATION, CONCURRENT MERCHANDISE SESSION LOUNGE, EDUCATION & OUTREACH SPEAKER READY COMMITTEE MEETING POSTER SESSION ROOM ROOM SOCIETY OF VERTEBRATE PALEONTOLOGY ABSTRACTS OF PAPERS SEVENTY-FIRST ANNUAL MEETING PARIS LAS VEGAS HOTEL LAS VEGAS, NV, USA NOVEMBER 2–5, 2011 HOST COMMITTEE Stephen Rowland, Co-Chair; Aubrey Bonde, Co-Chair; Joshua Bonde; David Elliott; Lee Hall; Jerry Harris; Andrew Milner; Eric Roberts EXECUTIVE COMMITTEE Philip Currie, President; Blaire Van Valkenburgh, Past President; Catherine Forster, Vice President; Christopher Bell, Secretary; Ted Vlamis, Treasurer; Julia Clarke, Member at Large; Kristina Curry Rogers, Member at Large; Lars Werdelin, Member at Large SYMPOSIUM CONVENORS Roger B.J. Benson, Richard J. Butler, Nadia B. Fröbisch, Hans C.E. Larsson, Mark A. Loewen, Philip D. Mannion, Jim I. Mead, Eric M. Roberts, Scott D. Sampson, Eric D. Scott, Kathleen Springer PROGRAM COMMITTEE Jonathan Bloch, Co-Chair; Anjali Goswami, Co-Chair; Jason Anderson; Paul Barrett; Brian Beatty; Kerin Claeson; Kristina Curry Rogers; Ted Daeschler; David Evans; David Fox; Nadia B. Fröbisch; Christian Kammerer; Johannes Müller; Emily Rayfield; William Sanders; Bruce Shockey; Mary Silcox; Michelle Stocker; Rebecca Terry November 2011—PROGRAM AND ABSTRACTS 1 Members and Friends of the Society of Vertebrate Paleontology, The Host Committee cordially welcomes you to the 71st Annual Meeting of the Society of Vertebrate Paleontology in Las Vegas. -

Full Issue 114
South African Journal of Science volume 114 number 5/6 Decolonising engineering in South Africa The Grootfontein aquifer: Governance of a hydro-social system Household food waste disposal in South Africa South African behavioural ecology research in a global perspective Volume 114 EDITOR-IN-CHIEF Number 5/6 John Butler-Adam Academy of Science of South Africa May/June 2018 MANAGING EDITOR Linda Fick Academy of Science of South Africa South African ONLINE PUBLISHING Journal of Science SYSTEMS ADMINISTRATOR Nadine Wubbeling Academy of Science of South Africa ONLINE PUBLISHING ADMINISTRATOR Sbonga Dlamini eISSN: 1996-7489 Academy of Science of South Africa ASSOCIATE EDITORS Nicolas Beukes Leader Department of Geology, University of Johannesburg The Fourth Industrial Revolution and education John Butler-Adam .................................................................................................................... 1 Chris Chimimba Department of Zoology and Entomology, University of Pretoria Book Review Towards human development friendly universities Linda Chisholm Merridy Wilson-Strydom .......................................................................................................... 2 Centre for Education Rights and Transformation, University of A new look at Old Fourlegs Johannesburg Brian W. van Wilgen ................................................................................................................. 4 Teresa Coutinho Department of Microbiology and Commentary Plant Pathology, University of Pretoria Decolonising