Introduction to Cladistic Analysis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Phylogenetic Trees and Cladograms Are Graphical Representations (Models) of Evolutionary History That Can Be Tested
AP Biology Lab/Cladograms and Phylogenetic Trees Name _______________________________ Relationship to the AP Biology Curriculum Framework Big Idea 1: The process of evolution drives the diversity and unity of life. Essential knowledge 1.B.2: Phylogenetic trees and cladograms are graphical representations (models) of evolutionary history that can be tested. Learning Objectives: LO 1.17 The student is able to pose scientific questions about a group of organisms whose relatedness is described by a phylogenetic tree or cladogram in order to (1) identify shared characteristics, (2) make inferences about the evolutionary history of the group, and (3) identify character data that could extend or improve the phylogenetic tree. LO 1.18 The student is able to evaluate evidence provided by a data set in conjunction with a phylogenetic tree or a simple cladogram to determine evolutionary history and speciation. LO 1.19 The student is able create a phylogenetic tree or simple cladogram that correctly represents evolutionary history and speciation from a provided data set. [Introduction] Cladistics is the study of evolutionary classification. Cladograms show evolutionary relationships among organisms. Comparative morphology investigates characteristics for homology and analogy to determine which organisms share a recent common ancestor. A cladogram will begin by grouping organisms based on a characteristics displayed by ALL the members of the group. Subsequently, the larger group will contain increasingly smaller groups that share the traits of the groups before them. However, they also exhibit distinct changes as the new species evolve. Further, molecular evidence from genes which rarely mutate can provide molecular clocks that tell us how long ago organisms diverged, unlocking the secrets of organisms that may have similar convergent morphology but do not share a recent common ancestor. -

An Introduction to Phylogenetic Analysis
This article reprinted from: Kosinski, R.J. 2006. An introduction to phylogenetic analysis. Pages 57-106, in Tested Studies for Laboratory Teaching, Volume 27 (M.A. O'Donnell, Editor). Proceedings of the 27th Workshop/Conference of the Association for Biology Laboratory Education (ABLE), 383 pages. Compilation copyright © 2006 by the Association for Biology Laboratory Education (ABLE) ISBN 1-890444-09-X All rights reserved. No part of this publication may be reproduced, stored in a retrieval system, or transmitted, in any form or by any means, electronic, mechanical, photocopying, recording, or otherwise, without the prior written permission of the copyright owner. Use solely at one’s own institution with no intent for profit is excluded from the preceding copyright restriction, unless otherwise noted on the copyright notice of the individual chapter in this volume. Proper credit to this publication must be included in your laboratory outline for each use; a sample citation is given above. Upon obtaining permission or with the “sole use at one’s own institution” exclusion, ABLE strongly encourages individuals to use the exercises in this proceedings volume in their teaching program. Although the laboratory exercises in this proceedings volume have been tested and due consideration has been given to safety, individuals performing these exercises must assume all responsibilities for risk. The Association for Biology Laboratory Education (ABLE) disclaims any liability with regards to safety in connection with the use of the exercises in this volume. The focus of ABLE is to improve the undergraduate biology laboratory experience by promoting the development and dissemination of interesting, innovative, and reliable laboratory exercises. -

A Phylogenetic Analysis of the Basal Ornithischia (Reptilia, Dinosauria)
A PHYLOGENETIC ANALYSIS OF THE BASAL ORNITHISCHIA (REPTILIA, DINOSAURIA) Marc Richard Spencer A Thesis Submitted to the Graduate College of Bowling Green State University in partial fulfillment of the requirements of the degree of MASTER OF SCIENCE December 2007 Committee: Margaret M. Yacobucci, Advisor Don C. Steinker Daniel M. Pavuk © 2007 Marc Richard Spencer All Rights Reserved iii ABSTRACT Margaret M. Yacobucci, Advisor The placement of Lesothosaurus diagnosticus and the Heterodontosauridae within the Ornithischia has been problematic. Historically, Lesothosaurus has been regarded as a basal ornithischian dinosaur, the sister taxon to the Genasauria. Recent phylogenetic analyses, however, have placed Lesothosaurus as a more derived ornithischian within the Genasauria. The Fabrosauridae, of which Lesothosaurus was considered a member, has never been phylogenetically corroborated and has been considered a paraphyletic assemblage. Prior to recent phylogenetic analyses, the problematic Heterodontosauridae was placed within the Ornithopoda as the sister taxon to the Euornithopoda. The heterodontosaurids have also been considered as the basal member of the Cerapoda (Ornithopoda + Marginocephalia), the sister taxon to the Marginocephalia, and as the sister taxon to the Genasauria. To reevaluate the placement of these taxa, along with other basal ornithischians and more derived subclades, a phylogenetic analysis of 19 taxonomic units, including two outgroup taxa, was performed. Analysis of 97 characters and their associated character states culled, modified, and/or rescored from published literature based on published descriptions, produced four most parsimonious trees. Consistency and retention indices were calculated and a bootstrap analysis was performed to determine the relative support for the resultant phylogeny. The Ornithischia was recovered with Pisanosaurus as its basalmost member. -

Interpreting Cladograms Notes
Interpreting Cladograms Notes INTERPRETING CLADOGRAMS BIG IDEA: PHYLOGENIES DEPICT ANCESTOR AND DESCENDENT RELATIONSHIPS AMONG ORGANISMS BASED ON HOMOLOGY THESE EVOLUTIONARY RELATIONSHIPS ARE REPRESENTED BY DIAGRAMS CALLED CLADOGRAMS (BRANCHING DIAGRAMS THAT ORGANIZE RELATIONSHIPS) What is a Cladogram ● A diagram which shows ● Is not an evolutionary ___________ among tree, ___________show organisms. how ancestors are related ● Lines branching off other to descendants or how lines. The lines can be much they have changed traced back to where they branch off. These branching off points represent a hypothetical ancestor. Parts of a Cladogram Reading Cladograms ● Read like a family tree: show ________of shared ancestry between lineages. • When an ancestral lineage______: speciation is indicated due to the “arrival” of some new trait. Each lineage has unique ____to itself alone and traits that are shared with other lineages. each lineage has _______that are unique to that lineage and ancestors that are shared with other lineages — common ancestors. Quick Question #1 ●What is a_________? ● A group that includes a common ancestor and all the descendants (living and extinct) of that ancestor. Reading Cladogram: Identifying Clades ● Using a cladogram, it is easy to tell if a group of lineages forms a clade. ● Imagine clipping a single branch off the phylogeny ● all of the organisms on that pruned branch make up a clade Quick Question #2 ● Looking at the image to the right: ● Is the green box a clade? ● The blue? ● The pink? ● The orange? Reading Cladograms: Clades ● Clades are nested within one another ● they form a nested hierarchy. ● A clade may include many thousands of species or just a few. -

Conceptual Issues in Phylogeny, Taxonomy, and Nomenclature
Contributions to Zoology, 66 (1) 3-41 (1996) SPB Academic Publishing bv, Amsterdam Conceptual issues in phylogeny, taxonomy, and nomenclature Alexandr P. Rasnitsyn Paleontological Institute, Russian Academy ofSciences, Profsoyuznaya Street 123, J17647 Moscow, Russia Keywords: Phylogeny, taxonomy, phenetics, cladistics, phylistics, principles of nomenclature, type concept, paleoentomology, Xyelidae (Vespida) Abstract On compare les trois approches taxonomiques principales développées jusqu’à présent, à savoir la phénétique, la cladis- tique et la phylistique (= systématique évolutionnaire). Ce Phylogenetic hypotheses are designed and tested (usually in dernier terme s’applique à une approche qui essaie de manière implicit form) on the basis ofa set ofpresumptions, that is, of à les traits fondamentaux de la taxonomic statements explicite représenter describing a certain order of things in nature. These traditionnelle en de leur et particulier son usage preuves ayant statements are to be accepted as such, no matter whatever source en même temps dans la similitude et dans les relations de evidence for them exists, but only in the absence ofreasonably parenté des taxons en question. L’approche phylistique pré- sound evidence pleading against them. A set ofthe most current sente certains avantages dans la recherche de réponses aux phylogenetic presumptions is discussed, and a factual example problèmes fondamentaux de la taxonomie. ofa practical realization of the approach is presented. L’auteur considère la nomenclature A is made of the three -

Bioinfo 11 Phylogenetictree
BIO 390/CSCI 390/MATH 390 Bioinformatics II Programming Lecture 11 Phylogenetic Tree Instructor: Lei Qian Fisk University Phylogenetic Tree Applications • Study ancestor-descendant relationships (Evolutionary biology, adaption, genetic drift, selection, speciation, etc.) • Paleogenomics: inferring ancestral genomic information from extinct species (Comparing Chimpanzee, Neanderthal and Human DNA) • Origins of epidemics (Comparing, at the molecular level, various virus strains) • Drug design: specially targeting groups of organisms (Efficient enumeration of phylogenetically informative substrings) • Forensic (Relationships among HIV strains) • Linguistics (Languages tree divergence times) Phylogenetic Tree Illustrating success stories in phylogenetics (I) For roughly 100 years (more exactly, 1870-1985), scientists were unable to figure out which family the giant panda belongs to. Giant pandas look like bears, but have features that are unusual for bears but typical to raccoons: they do not hibernate, they do not roar, their male genitalia are small and backward-pointing. Anatomical features were the dominant criteria used to derive evolutionary relationships between species since Darwin till early 1960s. The evolutionary relationships derived from these relatively subjective observations were often inconclusive. Some of them were later proved incorrect. In 1985, Steven O’Brien and colleagues solved the giant panda classification problem using DNA sequences and phylogenetic algorithms. Phylogenetic Tree Phylogenetic Tree Illustrating success stories in phylogenetics (II) In 1994, a woman from Lafayette, Louisiana (USA), clamed that her ex-lover (who was a physician) injected her with HIV+ blood. Records showed that the physician had drawn blood from a HIV+ patient that day. But how to prove that the blood from that HIV+ patient ended up in the woman? Phylogenetic Tree HIV has a high mutation rate, which can be used to trace paths of transmission. -

Hemidactylium Scutatum): out of Appalachia and Into the Glacial Aftermath
RANGE-WIDE PHYLOGEOGRAPHY OF THE FOUR-TOED SALAMANDER (HEMIDACTYLIUM SCUTATUM): OUT OF APPALACHIA AND INTO THE GLACIAL AFTERMATH Timothy A. Herman A Thesis Submitted to the Graduate College of Bowling Green State University in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE August 2009 Committee: Juan Bouzat, Advisor Christopher Phillips Karen Root ii ABSTRACT Juan Bouzat, Advisor Due to its limited vagility, deep ancestry, and broad distribution, the four-toed salamander (Hemidactylium scutatum) is well suited to track biogeographic patterns across eastern North America. The range of the monotypic genus Hemidactylium is highly disjunct in its southern and western portions, and even within contiguous portions is highly localized around pockets of preferred nesting habitat. Over 330 Hemidactylium genetic samples from 79 field locations were collected and analyzed via mtDNA sequencing of the cytochrome oxidase 1 gene (co1). Phylogenetic analyses showed deep divergences at this marker (>10% between some haplotypes) and strong support for regional monophyletic clades with minimal overlap. Patterns of haplotype distribution suggest major river drainages, both ancient and modern, as boundaries to dispersal. Two distinct allopatric clades account for all sampling sites within glaciated areas of North America yet show differing patterns of recolonization. High levels of haplotype diversity were detected in the southern Appalachians, with several members of widely ranging clades represented in the region as well as other unique, endemic, and highly divergent lineages. Bayesian divergence time analyses estimated the common ancestor of all living Hemidactylium included in the study at roughly 8 million years ago, with the most basal splits in the species confined to the Blue Ridge Mountains. -

Molecular Phylogenetics and Evolution 55 (2010) 153–167
Molecular Phylogenetics and Evolution 55 (2010) 153–167 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Conservation phylogenetics of helodermatid lizards using multiple molecular markers and a supertree approach Michael E. Douglas a,*, Marlis R. Douglas a, Gordon W. Schuett b, Daniel D. Beck c, Brian K. Sullivan d a Illinois Natural History Survey, Institute for Natural Resource Sustainability, University of Illinois, Champaign, IL 61820, USA b Department of Biology and Center for Behavioral Neuroscience, Georgia State University, Atlanta, GA 30303-3088, USA c Department of Biological Sciences, Central Washington University, Ellensburg, WA 98926, USA d Division of Mathematics & Natural Sciences, Arizona State University, Phoenix, AZ 85069, USA article info abstract Article history: We analyzed both mitochondrial (MT-) and nuclear (N) DNAs in a conservation phylogenetic framework to Received 30 June 2009 examine deep and shallow histories of the Beaded Lizard (Heloderma horridum) and Gila Monster (H. Revised 6 December 2009 suspectum) throughout their geographic ranges in North and Central America. Both MTDNA and intron Accepted 7 December 2009 markers clearly partitioned each species. One intron and MTDNA further subdivided H. horridum into its Available online 16 December 2009 four recognized subspecies (H. n. alvarezi, charlesbogerti, exasperatum, and horridum). However, the two subspecies of H. suspectum (H. s. suspectum and H. s. cinctum) were undefined. A supertree approach sus- Keywords: tained these relationships. Overall, the Helodermatidae is reaffirmed as an ancient and conserved group. Anguimorpha Its most recent common ancestor (MRCA) was Lower Eocene [35.4 million years ago (mya)], with a 25 ATPase Enolase my period of stasis before the MRCA of H. -

Supertree Construction: Opportunities and Challenges
Supertree Construction: Opportunities and Challenges Tandy Warnow Abstract Supertree construction is the process by which a set of phylogenetic trees, each on a subset of the overall set S of species, is combined into a tree on the full set S. The traditional use of supertree methods is the assembly of a large species tree from previously computed smaller species trees; however, supertree methods are also used to address large-scale tree estimation using divide-and-conquer (i.e., a dataset is divided into overlapping subsets, trees are constructed on the subsets, and then combined using the supertree method). Because most supertree methods are heuristics for NP-hard optimization problems, the use of supertree estimation on large datasets is challenging, both in terms of scalability and accuracy. In this chap- ter, we describe the current state of the art in supertree construction and the use of supertree methods in divide-and-conquer strategies. Finally, we identify directions where future research could lead to improved supertree methods. Key words: supertrees, phylogenetics, species trees, divide-and-conquer, incom- plete lineage sorting, Tree of Life 1 Introduction The estimation of phylogenetic trees, whether gene trees (that is, the phylogeny re- lating the sequences found at a specific locus in different species) or species trees, is a precursor to many downstream analyses, including protein structure and func- arXiv:1805.03530v1 [q-bio.PE] 9 May 2018 tion prediction, the detection of positive selection, co-evolution, human population genetics, etc. (see [58] for just some of these applications). Indeed, as Dobzhanksy said, “Nothing in biology makes sense except in the light of evolution” [44]. -

Phylogenetic Analysis
Phylogenetic Analysis Aristotle • Through classification, one might discover the essence and purpose of species. Nelson & Platnick (1981) Systematics and Biogeography Carl Linnaeus • Swedish botanist (1700s) • Listed all known species • Developed scheme of classification to discover the plan of the Creator 1 Linnaeus’ Main Contributions 1) Hierarchical classification scheme Kingdom: Phylum: Class: Order: Family: Genus: Species 2) Binomial nomenclature Before Linnaeus physalis amno ramosissime ramis angulosis glabris foliis dentoserratis After Linnaeus Physalis angulata (aka Cutleaf groundcherry) 3) Originated the practice of using the ♂ - (shield and arrow) Mars and ♀ - (hand mirror) Venus glyphs as the symbol for male and female. Charles Darwin • Species evolved from common ancestors. • Concept of closely related species being more recently diverged from a common ancestor. Therefore taxonomy might actually represent phylogeny! The phylogeny and classification of life a proposed by Haeckel (1866). 2 Trees - Rooted and Unrooted 3 Trees - Rooted and Unrooted ABCDEFGHIJ A BCDEH I J F G ROOT ROOT D E ROOT A F B H J G C I 4 Monophyletic: A group composed of a collection of organisms, including the most recent common ancestor of all those organisms and all the descendants of that most recent common ancestor. A monophyletic taxon is also called a clade. Paraphyletic: A group composed of a collection of organisms, including the most recent common ancestor of all those organisms. Unlike a monophyletic group, a paraphyletic group does not include all the descendants of the most recent common ancestor. Polyphyletic: A group composed of a collection of organisms in which the most recent common ancestor of all the included organisms is not included, usually because the common ancestor lacks the characteristics of the group. -
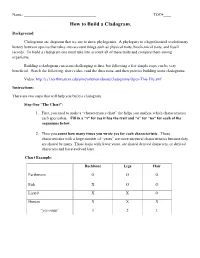
How to Build a Cladogram
Name: _____________________________________ TOC#____ How to Build a Cladogram. Background: Cladograms are diagrams that we use to show phylogenies. A phylogeny is a hypothesized evolutionary history between species that takes into account things such as physical traits, biochemical traits, and fossil records. To build a cladogram one must take into account all of these traits and compare them among organisms. Building a cladogram can seem challenging at first, but following a few simple steps can be very beneficial. Watch the following, short video, read the directions, and then practice building some cladograms. Video: http://ccl.northwestern.edu/simevolution/obonu/cladograms/Open-This-File.swf Instructions: There are two steps that will help you build a cladogram. Step One “The Chart”: 1. First, you need to make a “characteristics chart” the helps you analyze which characteristics each specieshas. Fill in a “x” for yes it has the trait and “o” for “no” for each of the organisms below. 2. Then you count how many times you wrote yes for each characteristic. Those characteristics with a large number of “yeses” are more ancestral characteristics because they are shared by many. Those traits with fewer yeses, are shared derived characters, or derived characters and have evolved later. Chart Example: Backbone Legs Hair Earthworm O O O Fish X O O Lizard X X O Human X X X “yes count” 3 2 1 Step Two “The Venn Diagram”: This step will help you to learn to build Cladograms, but once you figure it out, you may not always need to do this step. -

Introduction to Phylogenetics Workshop on Molecular Evolution 2018 Marine Biological Lab, Woods Hole, MA
Introduction to Phylogenetics Workshop on Molecular Evolution 2018 Marine Biological Lab, Woods Hole, MA. USA Mark T. Holder University of Kansas Outline 1. phylogenetics is crucial for comparative biology 2. tree terminology 3. why phylogenetics is difficult 4. parsimony 5. distance-based methods 6. theoretical basis of multiple sequence alignment Part #1: phylogenetics is crucial for biology Species Habitat Photoprotection 1 terrestrial xanthophyll 2 terrestrial xanthophyll 3 terrestrial xanthophyll 4 terrestrial xanthophyll 5 terrestrial xanthophyll 6 aquatic none 7 aquatic none 8 aquatic none 9 aquatic none 10 aquatic none slides by Paul Lewis Phylogeny reveals the events that generate the pattern 1 pair of changes. 5 pairs of changes. Coincidence? Much more convincing Many evolutionary questions require a phylogeny Determining whether a trait tends to be lost more often than • gained, or vice versa Estimating divergence times (Tracy Heath Sunday + next • Saturday) Distinguishing homology from analogy • Inferring parts of a gene under strong positive selection (Joe • Bielawski and Belinda Chang next Monday) Part 2: Tree terminology A B C D E terminal node (or leaf, degree 1) interior node (or vertex, degree 3+) split (bipartition) also written AB|CDE or portrayed **--- branch (edge) root node of tree (de gree 2) Monophyletic groups (\clades"): the basis of phylogenetic classification black state = a synapomorphy white state = a plesiomorphy Paraphyletic Polyphyletic grey state is an autapomorphy (images from Wikipedia) Branch rotation does not matter ACEBFDDAFBEC Rooted vs unrooted trees ingroup: the focal taxa outgroup: the taxa that are more distantly related. Assuming that the ingroup is monophyletic with respect to the outgroup can root a tree.