Discrete Probability Distributions Geometric and Negative Binomial Illustrated by Mitochondrial Eve and Cancer Driv
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Discrete Probability Distributions Uniform Distribution Bernoulli
Discrete Probability Distributions Uniform Distribution Experiment obeys: all outcomes equally probable Random variable: outcome Probability distribution: if k is the number of possible outcomes, 1 if x is a possible outcome p(x)= k ( 0 otherwise Example: tossing a fair die (k = 6) Bernoulli Distribution Experiment obeys: (1) a single trial with two possible outcomes (success and failure) (2) P trial is successful = p Random variable: number of successful trials (zero or one) Probability distribution: p(x)= px(1 − p)n−x Mean and variance: µ = p, σ2 = p(1 − p) Example: tossing a fair coin once Binomial Distribution Experiment obeys: (1) n repeated trials (2) each trial has two possible outcomes (success and failure) (3) P ith trial is successful = p for all i (4) the trials are independent Random variable: number of successful trials n x n−x Probability distribution: b(x; n,p)= x p (1 − p) Mean and variance: µ = np, σ2 = np(1 − p) Example: tossing a fair coin n times Approximations: (1) b(x; n,p) ≈ p(x; λ = pn) if p ≪ 1, x ≪ n (Poisson approximation) (2) b(x; n,p) ≈ n(x; µ = pn,σ = np(1 − p) ) if np ≫ 1, n(1 − p) ≫ 1 (Normal approximation) p Geometric Distribution Experiment obeys: (1) indeterminate number of repeated trials (2) each trial has two possible outcomes (success and failure) (3) P ith trial is successful = p for all i (4) the trials are independent Random variable: trial number of first successful trial Probability distribution: p(x)= p(1 − p)x−1 1 2 1−p Mean and variance: µ = p , σ = p2 Example: repeated attempts to start -

Chapter 5 Sections
Chapter 5 Chapter 5 sections Discrete univariate distributions: 5.2 Bernoulli and Binomial distributions Just skim 5.3 Hypergeometric distributions 5.4 Poisson distributions Just skim 5.5 Negative Binomial distributions Continuous univariate distributions: 5.6 Normal distributions 5.7 Gamma distributions Just skim 5.8 Beta distributions Multivariate distributions Just skim 5.9 Multinomial distributions 5.10 Bivariate normal distributions 1 / 43 Chapter 5 5.1 Introduction Families of distributions How: Parameter and Parameter space pf /pdf and cdf - new notation: f (xj parameters ) Mean, variance and the m.g.f. (t) Features, connections to other distributions, approximation Reasoning behind a distribution Why: Natural justification for certain experiments A model for the uncertainty in an experiment All models are wrong, but some are useful – George Box 2 / 43 Chapter 5 5.2 Bernoulli and Binomial distributions Bernoulli distributions Def: Bernoulli distributions – Bernoulli(p) A r.v. X has the Bernoulli distribution with parameter p if P(X = 1) = p and P(X = 0) = 1 − p. The pf of X is px (1 − p)1−x for x = 0; 1 f (xjp) = 0 otherwise Parameter space: p 2 [0; 1] In an experiment with only two possible outcomes, “success” and “failure”, let X = number successes. Then X ∼ Bernoulli(p) where p is the probability of success. E(X) = p, Var(X) = p(1 − p) and (t) = E(etX ) = pet + (1 − p) 8 < 0 for x < 0 The cdf is F(xjp) = 1 − p for 0 ≤ x < 1 : 1 for x ≥ 1 3 / 43 Chapter 5 5.2 Bernoulli and Binomial distributions Binomial distributions Def: Binomial distributions – Binomial(n; p) A r.v. -

(Introduction to Probability at an Advanced Level) - All Lecture Notes
Fall 2018 Statistics 201A (Introduction to Probability at an advanced level) - All Lecture Notes Aditya Guntuboyina August 15, 2020 Contents 0.1 Sample spaces, Events, Probability.................................5 0.2 Conditional Probability and Independence.............................6 0.3 Random Variables..........................................7 1 Random Variables, Expectation and Variance8 1.1 Expectations of Random Variables.................................9 1.2 Variance................................................ 10 2 Independence of Random Variables 11 3 Common Distributions 11 3.1 Ber(p) Distribution......................................... 11 3.2 Bin(n; p) Distribution........................................ 11 3.3 Poisson Distribution......................................... 12 4 Covariance, Correlation and Regression 14 5 Correlation and Regression 16 6 Back to Common Distributions 16 6.1 Geometric Distribution........................................ 16 6.2 Negative Binomial Distribution................................... 17 7 Continuous Distributions 17 7.1 Normal or Gaussian Distribution.................................. 17 1 7.2 Uniform Distribution......................................... 18 7.3 The Exponential Density...................................... 18 7.4 The Gamma Density......................................... 18 8 Variable Transformations 19 9 Distribution Functions and the Quantile Transform 20 10 Joint Densities 22 11 Joint Densities under Transformations 23 11.1 Detour to Convolutions...................................... -

Lecture Notes #17: Some Important Distributions and Coupon Collecting
EECS 70 Discrete Mathematics and Probability Theory Spring 2014 Anant Sahai Note 17 Some Important Distributions In this note we will introduce three important probability distributions that are widely used to model real- world phenomena. The first of these which we already learned about in the last Lecture Note is the binomial distribution Bin(n; p). This is the distribution of the number of Heads, Sn, in n tosses of a biased coin with n k n−k probability p to be Heads. As we saw, P[Sn = k] = k p (1 − p) . E[Sn] = np, Var[Sn] = np(1 − p) and p s(Sn) = np(1 − p). Geometric Distribution Question: A biased coin with Heads probability p is tossed repeatedly until the first Head appears. What is the distribution and the expected number of tosses? As always, our first step in answering the question must be to define the sample space W. A moment’s thought tells us that W = fH;TH;TTH;TTTH;:::g; i.e., W consists of all sequences over the alphabet fH;Tg that end with H and contain no other H’s. This is our first example of an infinite sample space (though it is still discrete). What is the probability of a sample point, say w = TTH? Since successive coin tosses are independent (this is implicit in the statement of the problem), we have Pr[TTH] = (1 − p) × (1 − p) × p = (1 − p)2 p: And generally, for any sequence w 2 W of length i, we have Pr[w] = (1 − p)i−1 p. -

Lecture 8: Geometric and Binomial Distributions
Recap Dangling thread from last time From OpenIntro quiz 2: Lecture 8: Geometric and Binomial distributions Statistics 101 Mine C¸etinkaya-Rundel P(took j not valuable) September 22, 2011 P(took and not valuable) = P(not valuable) 0:1403 = 0:1403 + 0:5621 = 0:1997 Statistics 101 (Mine C¸etinkaya-Rundel) L8: Geometric and Binomial September 22, 2011 1 / 27 Recap Geometric distribution Bernoulli distribution Milgram experiment Clicker question (graded) Stanley Milgram, a Yale University psychologist, conducted a series of Which of the following is false? experiments on obedience to authority starting in 1963. Experimenter (E) orders the teacher (a) The Z score for the mean of a distribution of any shape is 0. (T), the subject of the experiment, to (b) Half the observations in a distribution of any shape have positive give severe electric shocks to a Z scores. learner (L) each time the learner (c) The median of a right skewed distribution has a negative Z score. answers a question incorrectly. (d) Majority of the values in a left skewed distribution have positive Z The learner is actually an actor, and scores. the electric shocks are not real, but a prerecorded sound is played each time the teacher administers an electric shock. Statistics 101 (Mine C¸etinkaya-Rundel) L8: Geometric and Binomial September 22, 2011 2 / 27 Statistics 101 (Mine C¸etinkaya-Rundel) L8: Geometric and Binomial September 22, 2011 3 / 27 Geometric distribution Bernoulli distribution Geometric distribution Bernoulli distribution Milgram experiment (cont.) Bernouilli random variables These experiments measured the willingness of study Each person in Milgram’s experiment can be thought of as a trial. -
Discrete Distributions: Empirical, Bernoulli, Binomial, Poisson
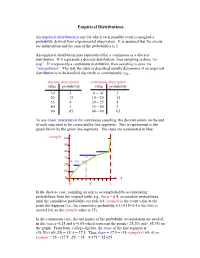
Empirical Distributions An empirical distribution is one for which each possible event is assigned a probability derived from experimental observation. It is assumed that the events are independent and the sum of the probabilities is 1. An empirical distribution may represent either a continuous or a discrete distribution. If it represents a discrete distribution, then sampling is done “on step”. If it represents a continuous distribution, then sampling is done via “interpolation”. The way the table is described usually determines if an empirical distribution is to be handled discretely or continuously; e.g., discrete description continuous description value probability value probability 10 .1 0 – 10- .1 20 .15 10 – 20- .15 35 .4 20 – 35- .4 40 .3 35 – 40- .3 60 .05 40 – 60- .05 To use linear interpolation for continuous sampling, the discrete points on the end of each step need to be connected by line segments. This is represented in the graph below by the green line segments. The steps are represented in blue: rsample 60 50 40 30 20 10 0 x 0 .5 1 In the discrete case, sampling on step is accomplished by accumulating probabilities from the original table; e.g., for x = 0.4, accumulate probabilities until the cumulative probability exceeds 0.4; rsample is the event value at the point this happens (i.e., the cumulative probability 0.1+0.15+0.4 is the first to exceed 0.4, so the rsample value is 35). In the continuous case, the end points of the probability accumulation are needed, in this case x=0.25 and x=0.65 which represent the points (.25,20) and (.65,35) on the graph. -

Some Continuous and Discrete Distributions Table of Contents I
Some continuous and discrete distributions Table of contents I. Continuous distributions and transformation rules. A. Standard uniform distribution U[0; 1]. B. Uniform distribution U[a; b]. C. Standard normal distribution N(0; 1). D. Normal distribution N(¹; ). E. Standard exponential distribution. F. Exponential distribution with mean ¸. G. Standard Gamma distribution ¡(r; 1). H. Gamma distribution ¡(r; ¸). II. Discrete distributions and transformation rules. A. Bernoulli random variables. B. Binomial distribution. C. Poisson distribution. D. Geometric distribution. E. Negative binomial distribution. F. Hypergeometric distribution. 1 Continuous distributions. Each continuous distribution has a \standard" version and a more general rescaled version. The transformation from one to the other is always of the form Y = aX + b, with a > 0, and the resulting identities: y b fX ¡ f (y) = a (1) Y ³a ´ y b FY (y) = FX ¡ (2) Ã a ! E(Y ) = aE(X) + b (3) V ar(Y ) = a2V ar(X) (4) bt MY (t) = e MX (at) (5) 1 1.1 Standard uniform U[0; 1] This distribution is \pick a random number between 0 and 1". 1 if 0 < x < 1 fX (x) = ½ 0 otherwise 0 if x 0 · FX (x) = x if 0 x 1 8 · · < 1 if x 1 ¸ E(X) = 1:=2 V ar(X) = 1=12 et 1 M (t) = ¡ X t 1.2 Uniform U[a; b] This distribution is \pick a random number between a and b". To get a random number between a and b, take a random number between 0 and 1, multiply it by b a, and add a. The properties of this random variable are obtained by applying¡ rules (1{5) to the previous subsection. -

Probability Distributions
Probability Distributions February 4, 2021 1 / 31 Deterministic vs Stochastic Models Until now, we have studied only deterministic models, in which future states are entirely specified by the current state of the system. In the real world, however, chance plays a major role in the dynamics of a population. Lightning may strike an individual. A fire may decimate a population. Individuals may fail to reproduce or produce a bonanza crop of offspring. New beneficial mutations may, by happenstance, occur in individuals that leave no children. Drought and famine may occur, or rains and excess. Probabilistic models include chance events and outcomes and can lead to results that differ from purely deterministic models. 2 / 31 Probability Distributions In this section, we will introduce the concept of a random variable and examine its probability distribution, expectation, and variance. Consider an event or series of events with a variety of possible outcomes. We define a random variable that represents a particular outcome of these events. Random variables are generally denoted by capital letters, e.g., X , Y , Z. For instance, toss a coin twice and record the number of heads. The number of heads is a random variable and it may take on three values: X = 0 (no heads), X = 1 (one head), and X = 2 (two heads). First, we will consider random variables, as in this example, which have only a discrete set of possible outcomes (e.g., 0, 1, 2). 3 / 31 Probability Distributions In many cases of interest, one can specify the probability distribution that a random variable will follow. -

A Compound Class of Geometric and Lifetimes Distributions
Send Orders of Reprints at [email protected] The Open Statistics and Probability Journal, 2013, 5, 1-5 1 Open Access A Compound Class of Geometric and Lifetimes Distributions Said Hofan Alkarni* Department of Quantitative Analysis, King Saud University, Riyadh, Saudi Arabia Abstract: A new lifetime class with decreasing failure rateis introduced by compounding truncated Geometric distribution and any proper continuous lifetime distribution. The properties of the proposed class are discussed, including a formal proof of its probability density function, distribution function and explicit algebraic formulae for its reliability and failure rate functions. A simple EM-type algorithm for iteratively computing maximum likelihood estimates is presented. A for- mal equation for Fisher information matrix is derived in order to obtaining the asymptotic covariance matrix. This new class of distributions generalizes several distributions which have been introduced and studied in the literature. Keywords: Lifetime distributions, decreasing failure rate, Geometric distribution. 1. INTRODUCTION and exponential with DFR was introduced by [7]. The expo- nential-Poisson (EP) distribution was proposed by [8] and The study of life time organisms, devices, structures, ma- generalized [9] using Wiebull distribution and the exponential- terials, etc., is of major importance in the biological and en- logarithmic distribution discussed by [10]. A two-parameter gineering sciences. A substantial part of such study is de- distribution family with decreasing -

Geometric, Negative Binomial Distributions
Geometric Distribution We will discuss two distributions in this section, both which have two parameterizations, so we must be careful. Suppose you roll a single die until you roll a 6. Let us consider \rolling a 6" a \success" and \not rolling a 6" a \failure". Our goal is to determine a distribution for the total number of rolls necessary to roll a 6. However, we can also write a distribution for the number of failures that occur, rather than the number of total rolls. Counting the number of failures results in a simpler probability formula, so this is what is typically used. The situation above, regardless of which term you count (number of failures or number of rolls) is a geo- metric distribution. We will derive the probability formula for this distribution, because it will help us get a feel for deriving the formula for the next distribution as well. Let X = the number of failures before the first success - in this case, the number of non-six rolls prior to 1 1 5 rolling a 6. The probability of rolling a 6 is 6 and the probability of a non-6 is 1 − 6 = 6 . Then, 1 50 1 P (X = 0) = = · Since there are no failures, we immediately succeed. 6 6 6 5 1 51 1 P (X = 1) = · = · Here we fail once, and then succeed. 6 6 6 6 5 5 1 52 1 P (X = 2) = · · = · Here we fail twice, and then succeed. 6 6 6 6 6 5 5 5 1 53 1 P (X = 3) = · · · = · Here we fail three times in a row, and then succeed. -

Discrete Random Variables and Probability Distributions
Discrete Random 3 Variables and Probability Distributions Stat 4570/5570 Based on Devore’s book (Ed 8) Random Variables We can associate each single outcome of an experiment with a real number: We refer to the outcomes of such experiments as a “random variable”. Why is it called a “random variable”? 2 Random Variables Definition For a given sample space S of some experiment, a random variable (r.v.) is a rule that associates a number with each outcome in the sample space S. In mathematical language, a random variable is a “function” whose domain is the sample space and whose range is the set of real numbers: X : R S ! So, for any event s, we have X(s)=x is a real number. 3 Random Variables Notation! 1. Random variables - usually denoted by uppercase letters near the end of our alphabet (e.g. X, Y). 2. Particular value - now use lowercase letters, such as x, which correspond to the r.v. X. Birth weight example 4 Two Types of Random Variables A discrete random variable: Values constitute a finite or countably infinite set A continuous random variable: 1. Its set of possible values is the set of real numbers R, one interval, or a disjoint union of intervals on the real line (e.g., [0, 10] ∪ [20, 30]). 2. No one single value of the variable has positive probability, that is, P(X = c) = 0 for any possible value c. Only intervals have positive probabilities. 5 Probability Distributions for Discrete Random Variables Probabilities assigned to various outcomes in the sample space S, in turn, determine probabilities associated with the values of any particular random variable defined on S. -

A Spreadsheet-Literate Non-Statistician's Guide to the Beta
A Spreadsheet-Literate Non-Statistician’s Guide to the Beta-Geometric Model Peter S. Fader www.petefader.com Bruce G. S. Hardie† www.brucehardie.com December 2014 1 Introduction The beta-geometric (BG) distribution is a robust simple model for characterizing and forecasting the length of a customer’s relationship with a firm in a contractual setting. Fader and Hardie (2007a), hereafter FH, is a definitive reference, providing a detailed derivation of the key quantities of interest, as well as step-by-step details of how to implement the model in Excel. However the statistical concepts and notation used in that paper can be daunting for those analysts who do not have a strong statistics background, leading them to ignore the model when it should really be a basic tool in their analytics toolkit. With spreadsheet-literate non-statisticians in mind as the target audience, the objective of this note is to describe the logic of the BG model in a non-technical manner and show how to implement it in Excel. 2 Motivating Problem Consider a company with a subscription-based business model that acquired 1000 customers (on annual contracts) at the beginning of Year 1. Table 1 reports the pattern of renewals by this cohort over the subsequent four years.1 We would like to predict how many members of this cohort will still be customers of the firm in Years 6, 7, .... † c 2014 Peter S. Fader and Bruce G. S. Hardie. This document and the associated spreadsheet (BG intro.xlsx) can be found at <http://brucehardie.com/notes/032/>.