The Landscape of Somatic Genetic Alterations in Metaplastic Breast Carcinomas
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

ORIGINAL ARTICLE Flow Cytometric Protein Expression Profiling As a Systematic Approach for Developing Disease-Specific Assays
Leukemia (2006) 20, 2102–2110 & 2006 Nature Publishing Group All rights reserved 0887-6924/06 $30.00 www.nature.com/leu ORIGINAL ARTICLE Flow cytometric protein expression profiling as a systematic approach for developing disease-specific assays: identification of a chronic lymphocytic leukaemia-specific assay for use in rituximab-containing regimens AC Rawstron, R de Tute, AS Jack and P Hillmen Haematological Malignancy Diagnostic Service (HMDS), Leeds Teaching Hospitals, Leeds, UK Depletion of disease below the levels detected by sensitive sustained remissions only occur in patients achieving an MRD- minimal residual disease (MRD) assays is associated with negative complete response.12 Therefore MRD is increasingly prolonged survival in chronic lymphocytic leukaemia (CLL). being used as an end point for therapeutic trials, and several Flow cytometric MRD assays are now sufficiently sensitive and rapid to guide the duration of therapy in CLL, but generally rely studies are now using the assessment of MRD to define the on assessment of CD20 expression, which cannot be accurately duration of therapy. measured during and after therapeutic approaches containing Approaches using allele-specific oligonucleotide polymerase rituximab. The aim of this study was to use analytical software chain reaction (ASO-PCR) to the immunoglobulin gene of the developed for microarray analysis to provide a systematic B-CLL cell are generally accepted to show the highest sensitivity approach for MRD flow assay development. Samples from CLL for MRD detection. However, more recent four-colour ap- patients (n ¼ 49), normal controls (n ¼ 21) and other B-lympho- proaches show sensitivities nearing that of ASO-PCR6,11,13 with proliferative disorders (n ¼ 12) were assessed with a panel of 66 antibodies. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Flow Reagents Single Color Antibodies CD Chart
CD CHART CD N° Alternative Name CD N° Alternative Name CD N° Alternative Name Beckman Coulter Clone Beckman Coulter Clone Beckman Coulter Clone T Cells B Cells Granulocytes NK Cells Macrophages/Monocytes Platelets Erythrocytes Stem Cells Dendritic Cells Endothelial Cells Epithelial Cells T Cells B Cells Granulocytes NK Cells Macrophages/Monocytes Platelets Erythrocytes Stem Cells Dendritic Cells Endothelial Cells Epithelial Cells T Cells B Cells Granulocytes NK Cells Macrophages/Monocytes Platelets Erythrocytes Stem Cells Dendritic Cells Endothelial Cells Epithelial Cells CD1a T6, R4, HTA1 Act p n n p n n S l CD99 MIC2 gene product, E2 p p p CD223 LAG-3 (Lymphocyte activation gene 3) Act n Act p n CD1b R1 Act p n n p n n S CD99R restricted CD99 p p CD224 GGT (γ-glutamyl transferase) p p p p p p CD1c R7, M241 Act S n n p n n S l CD100 SEMA4D (semaphorin 4D) p Low p p p n n CD225 Leu13, interferon induced transmembrane protein 1 (IFITM1). p p p p p CD1d R3 Act S n n Low n n S Intest CD101 V7, P126 Act n p n p n n p CD226 DNAM-1, PTA-1 Act n Act Act Act n p n CD1e R2 n n n n S CD102 ICAM-2 (intercellular adhesion molecule-2) p p n p Folli p CD227 MUC1, mucin 1, episialin, PUM, PEM, EMA, DF3, H23 Act p CD2 T11; Tp50; sheep red blood cell (SRBC) receptor; LFA-2 p S n p n n l CD103 HML-1 (human mucosal lymphocytes antigen 1), integrin aE chain S n n n n n n n l CD228 Melanotransferrin (MT), p97 p p CD3 T3, CD3 complex p n n n n n n n n n l CD104 integrin b4 chain; TSP-1180 n n n n n n n p p CD229 Ly9, T-lymphocyte surface antigen p p n p n -

CD Markers Are Routinely Used for the Immunophenotyping of Cells
ptglab.com 1 CD MARKER ANTIBODIES www.ptglab.com Introduction The cluster of differentiation (abbreviated as CD) is a protocol used for the identification and investigation of cell surface molecules. So-called CD markers are routinely used for the immunophenotyping of cells. Despite this use, they are not limited to roles in the immune system and perform a variety of roles in cell differentiation, adhesion, migration, blood clotting, gamete fertilization, amino acid transport and apoptosis, among many others. As such, Proteintech’s mini catalog featuring its antibodies targeting CD markers is applicable to a wide range of research disciplines. PRODUCT FOCUS PECAM1 Platelet endothelial cell adhesion of blood vessels – making up a large portion molecule-1 (PECAM1), also known as cluster of its intracellular junctions. PECAM-1 is also CD Number of differentiation 31 (CD31), is a member of present on the surface of hematopoietic the immunoglobulin gene superfamily of cell cells and immune cells including platelets, CD31 adhesion molecules. It is highly expressed monocytes, neutrophils, natural killer cells, on the surface of the endothelium – the thin megakaryocytes and some types of T-cell. Catalog Number layer of endothelial cells lining the interior 11256-1-AP Type Rabbit Polyclonal Applications ELISA, FC, IF, IHC, IP, WB 16 Publications Immunohistochemical of paraffin-embedded Figure 1: Immunofluorescence staining human hepatocirrhosis using PECAM1, CD31 of PECAM1 (11256-1-AP), Alexa 488 goat antibody (11265-1-AP) at a dilution of 1:50 anti-rabbit (green), and smooth muscle KD/KO Validated (40x objective). alpha-actin (red), courtesy of Nicola Smart. PECAM1: Customer Testimonial Nicola Smart, a cardiovascular researcher “As you can see [the immunostaining] is and a group leader at the University of extremely clean and specific [and] displays Oxford, has said of the PECAM1 antibody strong intercellular junction expression, (11265-1-AP) that it “worked beautifully as expected for a cell adhesion molecule.” on every occasion I’ve tried it.” Proteintech thanks Dr. -
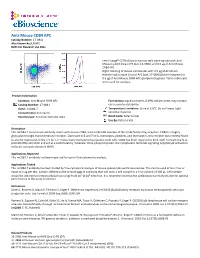
Anti-Mouse CD84 APC Catalog Number: 17‐0841 Also Known As:SLAMF5 RUO: for Research Use Only
Anti-Mouse CD84 APC Catalog Number: 17‐0841 Also Known As:SLAMF5 RUO: For Research Use Only Left: Lineagelo C57BL/6 bone marrow cells were stained with Anti‐ Mouse Ly‐6A/E (Sca‐1) PE (cat. 12‐5981) and 0.5 µg of Anti‐Mouse CD84 APC. Right: Staining of mouse splenocytes with 0.5 µg of Armenian Hamster IgG Isotype Control APC (cat. 17‐4888) (blue histogram) or 0.5 µg of Anti‐Mouse CD84 APC (purple histogram). Total viable cells were used for analysis. Product Information Contents: Anti‐Mouse CD84 APC Formulation: aqueous buffer, 0.09% sodium azide, may contain Catalog Number: 17‐0841 carrier protein/stabilizer Clone: mCD84.7 Temperature Limitation: Store at 2‐8°C. Do not freeze. Light Concentration: 0.2 mg/mL sensitive material. Host/Isotype: Armenian Hamster IgG1 Batch Code: Refer to Vial Use By: Refer to Vial Description This mCD84.7 monoclonal antibody reacts with mouse CD84, a 64‐ to 82‐kDa member of the SLAM family of Ig receptors. CD84 is a highly glycosylated single transmembrane receptor. Expressed in B and T cells, monocytes, platelets, and thymocytes, this receptor was recently found to also be expressed on Sca‐1+c‐kit+Lin‐ mouse bone marrow hematopoietic stem cells. CD84 has been reported to bind itself homophilically to promote IFNγ secretion and act as a costimulatory molecule. Once phosphorylated, the cytoplasmic tail binds signaling lymphocyte activation molecule‐associated protein (SAP). Applications Reported This mCD84.7 antibody has been reported for use in flow cytometric analysis. Applications Tested This mCD84.7 antibody has been tested by flow cytometric analysis of mouse splenocytes and bone marrow. -

Signalling Lymphocyte Activation Molecule Family Member 9 Is Found on Select Subsets of Antigen-Presenting Cells and Promotes Resistance to Salmonella Infection
IMMUNOLOGY ORIGINAL ARTICLE Signalling lymphocyte activation molecule family member 9 is found on select subsets of antigen-presenting cells and promotes resistance to Salmonella infection Timothy J. Wilson,1,2 Summary Simon Clare,3 Joseph Mikulin,1 Signalling lymphocyte activation molecule family member 9 (SLAMF9) is Christopher M. Johnson,4 an orphan receptor of the CD2/SLAM family of leucocyte surface pro- Katherine Harcourt,3 Paul A. teins. Examination of SLAMF9 expression and function indicates that Lyons,2,5 Gordon Dougan3,5 and SLAMF9 promotes inflammation by specialized subsets of antigen-pre- Kenneth G. C. Smith2,5 1 senting cells. Within healthy liver and circulating mouse peripheral blood Department of Microbiology, Miami + À low 2 mononuclear cells, SLAMF9 is expressed on CD11b , Ly6C , CD11c , University, Oxford, OH, USA, Department low + + of Medicine, Cambridge Institute for Medical F4/80 , MHC-II ,CX3CR1 mononuclear phagocytes as well as plasma- Research, University of Cambridge, cytoid dendritic cells. In addition, SLAMF9 can be found on peritoneal Cambridge, 3Wellcome Trust Sanger Institute, B1 cells and small (F4/80low), but not large (F4/80high), peritoneal macro- 4 Hinxton, Cambridge, MRC Laboratory of phages. Upon systemic challenge with Salmonella enterica Typhimurium, À/À Molecular Biology, Cambridge, and Slamf9 mice were impaired in their ability to clear the infection from 5Cambridge Institute for Therapeutic Immunology and Infectious Disease, the liver. In humans, SLAMF9 is up-regulated upon differentiation of Department of Medicine, University of monocytes into macrophages, and lipopolysaccharide stimulation of Cambridge, Cambridge, UK PMA-differentiated, SLAMF9 knockdown THP-1 cells showed an essential role of SLAMF9 in production of granulocyte–macrophage colony-stimu- doi:10.1111/imm.13169 lating factor, tumour necrosis factor-a, and interleukin-1b. -

The VE-Cadherin/Amotl2 Mechanosensory Pathway Suppresses Aortic In�Ammation and the Formation of Abdominal Aortic Aneurysms
The VE-cadherin/AmotL2 mechanosensory pathway suppresses aortic inammation and the formation of abdominal aortic aneurysms Yuanyuan Zhang Karolinska Institute Evelyn Hutterer Karolinska Institute Sara Hultin Karolinska Institute Otto Bergman Karolinska Institute Maria Forteza Karolinska Institute Zorana Andonovic Karolinska Institute Daniel Ketelhuth Karolinska University Hospital, Stockholm, Sweden Joy Roy Karolinska Institute Per Eriksson Karolinska Institute Lars Holmgren ( [email protected] ) Karolinska Institute Article Keywords: arterial endothelial cells (ECs), vascular disease, abdominal aortic aneurysms Posted Date: June 15th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-600069/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License The VE-cadherin/AmotL2 mechanosensory pathway suppresses aortic inflammation and the formation of abdominal aortic aneurysms Yuanyuan Zhang1, Evelyn Hutterer1, Sara Hultin1, Otto Bergman2, Maria J. Forteza2, Zorana Andonovic1, Daniel F.J. Ketelhuth2,3, Joy Roy4, Per Eriksson2 and Lars Holmgren1*. 1Department of Oncology-Pathology, BioClinicum, Karolinska Institutet, Stockholm, Sweden. 2Department of Medicine Solna, BioClinicum, Karolinska Institutet, Karolinska University Hospital, Stockholm, Sweden. 3Department of Cardiovascular and Renal Research, Institutet of Molecular Medicine, Univ. of Southern Denmark, Odense, Denmark 4Department of Molecular Medicine and Surgery, Karolinska Institutet, Karolinska University Hospital, Stockholm, -

That Provide Help for B Cells Their Role As Non-Th1/Th2 Effector Cells
T Follicular Helper Cells Express a Distinctive Transcriptional Profile, Reflecting Their Role as Non-Th1/Th2 Effector Cells That Provide Help for B Cells This information is current as of September 30, 2021. Tatyana Chtanova, Stuart G. Tangye, Rebecca Newton, Nita Frank, Martin R. Hodge, Michael S. Rolph and Charles R. Mackay J Immunol 2004; 173:68-78; ; doi: 10.4049/jimmunol.173.1.68 Downloaded from http://www.jimmunol.org/content/173/1/68 Supplementary http://www.jimmunol.org/content/suppl/2004/06/17/173.1.68.DC1 http://www.jimmunol.org/ Material References This article cites 63 articles, 32 of which you can access for free at: http://www.jimmunol.org/content/173/1/68.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision by guest on September 30, 2021 • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2004 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology T Follicular Helper Cells Express a Distinctive Transcriptional Profile, Reflecting Their Role as Non-Th1/Th2 Effector Cells That Provide Help for B Cells1 Tatyana Chtanova,*‡ Stuart G. -

CD84 Cell Surface Signaling Molecule an Emerging Biomarker
Clinical Immunology 204 (2019) 43–49 Contents lists available at ScienceDirect Clinical Immunology journal homepage: www.elsevier.com/locate/yclim Review Article CD84 cell surface signaling molecule: An emerging biomarker and target for cancer and autoimmune disorders T ⁎ Marta Cuencaa, , Jordi Sintesa, Árpád Lányib, Pablo Engela,c a Immunology Unit, Department of Biomedical Sciences, Faculty of Medicine, University of Barcelona, Barcelona, Spain b Department of Immunology, Faculty of Medicine, University of Debrecen, Debrecen, Hungary c Institut d'Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), Barcelona, Spain ARTICLE INFO ABSTRACT Keywords: CD84 (SLAMF5) is a member of the SLAM family of cell-surface immunoreceptors. Broadly expressed on most CD84 immune cell subsets, CD84 functions as a homophilic adhesion molecule, whose signaling can activate or inhibit SLAM family receptors leukocyte function depending on the cell type and its stage of activation or differentiation. CD84-mediated Germinal center (GC) signaling regulates diverse immunological processes, including T cell cytokine secretion, natural killer cell cy- Autophagy totoxicity, monocyte activation, autophagy, cognate T:B interactions, and B cell tolerance at the germinal center Disease biomarkers checkpoint. Recently, alterations in CD84 have been related to autoimmune and lymphoproliferative disorders. Autoimmune disorders fi Chronic lymphocytic leukemia (CLL) Speci c allelic variations in CD84 are associated with autoimmune diseases such as systemic lupus er- ythematosus and rheumatoid arthritis. In chronic lymphocytic leukemia, CD84 mediates intrinsic and stroma- induced survival of malignant cells. In this review, we describe our current understanding of the structure and function of CD84 and its potential role as a therapeutic target and biomarker in inflammatory autoimmune disorders and cancer. -

Studies on the Function and Regulation of CD84, GPVI and Orai2 in Genetically Modified Mice
Julius-Maximilians-Universität Würzburg Studies on the function and regulation of CD84, GPVI and Orai2 in genetically modified mice Untersuchungen zur Funktion und Regulation von CD84, GPVI und Orai2 in genetisch veränderten Mäusen Doctoral thesis for a doctoral degree at the Graduate School of Life Sciences Section Biomedicine submitted by Sebastian Hofmann from Coburg Würzburg, 2013 Submitted on: ………………………………………………………………………………… Office stamp Members of the Promotionskomitee: Chairperson: Prof. Dr. Manfred Gessler Primary Supervisor: Prof. Dr. Bernhard Nieswandt Supervisor (Second): PD Dr. Heike Hermanns Supervisor (Third): Prof. Dr. Guido Stoll Date of Public Defence: ……………………………………………………………………… Date of Receipt of Certificates: ……………………………………………………………... Summary Summary Platelet activation and aggregation at sites of vascular injury are essential processes to limit blood loss but they also contribute to arterial thrombosis, which can lead to myocardial infarction and stroke. Stable thrombus formation requires a series of events involving platelet receptors which contribute to adhesion, activation and aggregation of platelets. Regulation of receptor expression by (metallo-)proteinases has been described for several platelet receptors, but the molecular mechanisms are ill-defined. The signaling lymphocyte activation molecule (SLAM) family member CD84 is expressed in immune cells and platelets, however its role in platelet physiology was unclear. In this thesis, CD84 deficient mice were generated and analyzed. In well established in vitro and in vivo assays testing platelet function and thrombus formation, CD84 deficient mice displayed phenotypes indistinguishable from wild-type controls. It was concluded that CD84 in platelets does not function as modulator of thrombus formation, but rather has other functions. In line with this, in the second part of this thesis, a novel regulation mechanism for platelet CD84 was discovered and elucidated.