Polyploidy, Infraspecific Cytotype Variation, and Speciation in Goldenrods: the Cytogeography of Solidago Subsect
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Human Chimera: Legal Problems Arising from Individuals with Multiple Types of Dna
Seton Hall University eRepository @ Seton Hall Law School Student Scholarship Seton Hall Law 5-2-2014 The umH an Chimera: Legal Problems Arising From Individuals with Multiple Types of DNA Robert Russell Granzen Follow this and additional works at: https://scholarship.shu.edu/student_scholarship Recommended Citation Granzen, Robert Russell, "The umH an Chimera: Legal Problems Arising From Individuals with Multiple Types of DNA" (2014). Law School Student Scholarship. 485. https://scholarship.shu.edu/student_scholarship/485 THE HUMAN CHIMERA: LEGAL PROBLEMS ARISING FROM INDIVIDUALS WITH MULTIPLE TYPES OF DNA Robert Granzen INTRODUCTION Science continually changes, and with it our understanding of the human body. While some scientific developments are limited in scope, others have widespread effects. Scientists have just recently begun understanding the range of effects chimerism in humans can have. Chimerism, originally associated with hermaphrodites having both male and female sexual organs, is much more common than originally thought. As chimerism becomes more common, so do individuals with separate and distinct deoxyribonucleic acid (DNA) strands in their bodies. Most individuals are unaware of their chimeric genetic code and most will likely never know. Because of the inherent difficulty of testing for chimerism, many problems are presented to legal system. Part I of this article will begin with the history of human chimeras. The section will then describe the ways in which chimeras are formed. The section will discuss the most common form of chimerism in humans--fetal cell microchimerism (FMC). FMC occurs when cells are transferred from baby to mother or mother to baby via the umbilical cord.1 Studies have shown that mothers may keep cells from their children for years after giving birth.2 Moreover, cells can be exchanged between twins while inside the uterus.3 Secondly, the section will describe the process of embryo fusion, which can cause tetragametic chimerism. -

Natural Heritage Program List of Rare Plant Species of North Carolina 2016
Natural Heritage Program List of Rare Plant Species of North Carolina 2016 Revised February 24, 2017 Compiled by Laura Gadd Robinson, Botanist John T. Finnegan, Information Systems Manager North Carolina Natural Heritage Program N.C. Department of Natural and Cultural Resources Raleigh, NC 27699-1651 www.ncnhp.org C ur Alleghany rit Ashe Northampton Gates C uc Surry am k Stokes P d Rockingham Caswell Person Vance Warren a e P s n Hertford e qu Chowan r Granville q ot ui a Mountains Watauga Halifax m nk an Wilkes Yadkin s Mitchell Avery Forsyth Orange Guilford Franklin Bertie Alamance Durham Nash Yancey Alexander Madison Caldwell Davie Edgecombe Washington Tyrrell Iredell Martin Dare Burke Davidson Wake McDowell Randolph Chatham Wilson Buncombe Catawba Rowan Beaufort Haywood Pitt Swain Hyde Lee Lincoln Greene Rutherford Johnston Graham Henderson Jackson Cabarrus Montgomery Harnett Cleveland Wayne Polk Gaston Stanly Cherokee Macon Transylvania Lenoir Mecklenburg Moore Clay Pamlico Hoke Union d Cumberland Jones Anson on Sampson hm Duplin ic Craven Piedmont R nd tla Onslow Carteret co S Robeson Bladen Pender Sandhills Columbus New Hanover Tidewater Coastal Plain Brunswick THE COUNTIES AND PHYSIOGRAPHIC PROVINCES OF NORTH CAROLINA Natural Heritage Program List of Rare Plant Species of North Carolina 2016 Compiled by Laura Gadd Robinson, Botanist John T. Finnegan, Information Systems Manager North Carolina Natural Heritage Program N.C. Department of Natural and Cultural Resources Raleigh, NC 27699-1651 www.ncnhp.org This list is dynamic and is revised frequently as new data become available. New species are added to the list, and others are dropped from the list as appropriate. -

Association Between First-Trimester Intrauterine Hematoma and Twin Pregnancy Outcomes: a Retrospective Cohort Study
Association Between First-Trimester Intrauterine Hematoma and Twin Pregnancy Outcomes: A Retrospective Cohort Study Wanqing Ji Guangzhou Women and Children's Medical Center Jie Zheng Guangzhou Women and Children's Medical Center Weidong Li Guangzhou Women and Children's Medical Center Fang Guo Guangzhou Women and Children's Medical Center Bo Hou Third Aliated Hospital of Sun Yat-Sen University Ping He ( [email protected] ) Guangzhou Women and Children's Medical Center https://orcid.org/0000-0002-6551-8578 Research article Keywords: intrauterine hematoma, twin gestation, rst trimester, miscarriage, vanishing twin syndrome Posted Date: September 21st, 2020 DOI: https://doi.org/10.21203/rs.3.rs-75567/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Version of Record: A version of this preprint was published on January 11th, 2021. See the published version at https://doi.org/10.1186/s12884-020-03528-0. 1 Title page 2 Association between first-trimester intrauterine hematoma and twin pregnancy 3 outcomes: A retrospective cohort study 4 Wanqing Ji1,Ϯ , jie Zheng1,Ϯ, Weidong Li 2,Ϯ, Fang Guo1, , Ping He1,* Bo Hou3,* 5 1Department of Obstetrics, Guangzhou Women and Children’s Medical Center, Guangzhou Medical 6 University, Guangzhou, 510623,China. 7 2Department of Woman and Child Health Information Guangzhou Women and Children’s Medical 8 Center, Guangzhou Medical University, Guangzhou, 510623,China. 9 3Departments of Neurosurgery, The Third Affiliated Hospital, Sun Yat-sen University, Guangzhou, 10 Guangdong province, 510630, China.E-mail: [email protected] 11 *Correspondence address. Ping He, Guangzhou Women and Children’s Medical Center, Guangzhou 12 Medical University, Guangzhou, Guangdong province, 510623, China.Fax number: 86-20-8717 6790 13 E-mail: [email protected] 14 Bo Hou , Departments of Neurosurgery, The Third Affiliated Hospital, Sun Yat-sen University, 15 Guangzhou, Guangdong province, 510630, China. -

The Vascular Plants of Massachusetts
The Vascular Plants of Massachusetts: The Vascular Plants of Massachusetts: A County Checklist • First Revision Melissa Dow Cullina, Bryan Connolly, Bruce Sorrie and Paul Somers Somers Bruce Sorrie and Paul Connolly, Bryan Cullina, Melissa Dow Revision • First A County Checklist Plants of Massachusetts: Vascular The A County Checklist First Revision Melissa Dow Cullina, Bryan Connolly, Bruce Sorrie and Paul Somers Massachusetts Natural Heritage & Endangered Species Program Massachusetts Division of Fisheries and Wildlife Natural Heritage & Endangered Species Program The Natural Heritage & Endangered Species Program (NHESP), part of the Massachusetts Division of Fisheries and Wildlife, is one of the programs forming the Natural Heritage network. NHESP is responsible for the conservation and protection of hundreds of species that are not hunted, fished, trapped, or commercially harvested in the state. The Program's highest priority is protecting the 176 species of vertebrate and invertebrate animals and 259 species of native plants that are officially listed as Endangered, Threatened or of Special Concern in Massachusetts. Endangered species conservation in Massachusetts depends on you! A major source of funding for the protection of rare and endangered species comes from voluntary donations on state income tax forms. Contributions go to the Natural Heritage & Endangered Species Fund, which provides a portion of the operating budget for the Natural Heritage & Endangered Species Program. NHESP protects rare species through biological inventory, -

CDC Having Healthy Babies One at a Time
HAVING HEALTHY BABIES ONE AT A TIME Why are we worried about twin pregnancies? We know that you are ready to start or add to your family. You may be concerned about your chances of having a baby using in vitro fertilization (IVF) or how much cycles of IVF cost. These concerns are common and may lead you to think about transferring more than one embryo during your IVF procedure. However, transferring more than one embryo increases your chances of having twins or more. Twin pregnancy is risky for baby and mother, whether or not IVF is used. Some of these risks include: Almost 3 out of 5 Twin babies are more likely to be stillborn, twin babies are born preterm, or at less than experience neonatal death, have birth defects of 37 weeks of pregnancy. Twin babies are nearly the brain, heart, face, limbs, muscles, or digestive 6 times as likely to be born preterm as system, and have autism than single babies. single babies. Almost 1 out of 10 About 1 out of 4 women carrying twins gets pregnancy-related twin babies are admitted to the neonatal high blood pressure. Women carrying twins intensive care unit (NICU). Twin babies are are twice as likely to get pregnancy-related high more than 5 times as likely to be admitted to the blood pressure as women carrying single babies. NICU as single babies. Almost 1 out of 20 About 7 out of 1,000 women carrying twins gets gestational diabetes. twin babies have cerebral palsy. Twin babies are Women carrying twins are 1.5 times as likely to get more than 4 times as likely to have cerebral palsy gestational diabetes as women carrying as single babies. -

Natural Landscapes of Maine a Guide to Natural Communities and Ecosystems
Natural Landscapes of Maine A Guide to Natural Communities and Ecosystems by Susan Gawler and Andrew Cutko Natural Landscapes of Maine A Guide to Natural Communities and Ecosystems by Susan Gawler and Andrew Cutko Copyright © 2010 by the Maine Natural Areas Program, Maine Department of Conservation 93 State House Station, Augusta, Maine 04333-0093 All rights reserved. No part of this book may be reproduced or transmitted in any form or by any means, electronic or mechanical, including photocopying, recording, or by any information storage and retrieval system without written permission from the authors or the Maine Natural Areas Program, except for inclusion of brief quotations in a review. Illustrations and photographs are used with permission and are copyright by the contributors. Images cannot be reproduced without expressed written consent of the contributor. ISBN 0-615-34739-4 To cite this document: Gawler, S. and A. Cutko. 2010. Natural Landscapes of Maine: A Guide to Natural Communities and Ecosystems. Maine Natural Areas Program, Maine Department of Conservation, Augusta, Maine. Cover photo: Circumneutral Riverside Seep on the St. John River, Maine Printed and bound in Maine using recycled, chlorine-free paper Contents Page Acknowledgements ..................................................................................... 3 Foreword ..................................................................................................... 4 Introduction ............................................................................................... -
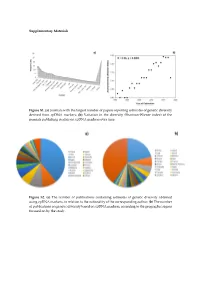
(A) Journals with the Largest Number of Papers Reporting Estimates Of
Supplementary Materials Figure S1. (a) Journals with the largest number of papers reporting estimates of genetic diversity derived from cpDNA markers; (b) Variation in the diversity (Shannon-Wiener index) of the journals publishing studies on cpDNA markers over time. Figure S2. (a) The number of publications containing estimates of genetic diversity obtained using cpDNA markers, in relation to the nationality of the corresponding author; (b) The number of publications on genetic diversity based on cpDNA markers, according to the geographic region focused on by the study. Figure S3. Classification of the angiosperm species investigated in the papers that analyzed genetic diversity using cpDNA markers: (a) Life mode; (b) Habitat specialization; (c) Geographic distribution; (d) Reproductive cycle; (e) Type of flower, and (f) Type of pollinator. Table S1. Plant species identified in the publications containing estimates of genetic diversity obtained from the use of cpDNA sequences as molecular markers. Group Family Species Algae Gigartinaceae Mazzaella laminarioides Angiospermae Typhaceae Typha laxmannii Angiospermae Typhaceae Typha orientalis Angiospermae Typhaceae Typha angustifolia Angiospermae Typhaceae Typha latifolia Angiospermae Araliaceae Eleutherococcus sessiliflowerus Angiospermae Polygonaceae Atraphaxis bracteata Angiospermae Plumbaginaceae Armeria pungens Angiospermae Aristolochiaceae Aristolochia kaempferi Angiospermae Polygonaceae Atraphaxis compacta Angiospermae Apocynaceae Lagochilus macrodontus Angiospermae Polygonaceae Atraphaxis -

Literature Cited
Literature Cited Robert W. Kiger, Editor This is a consolidated list of all works cited in volumes 19, 20, and 21, whether as selected references, in text, or in nomenclatural contexts. In citations of articles, both here and in the taxonomic treatments, and also in nomenclatural citations, the titles of serials are rendered in the forms recommended in G. D. R. Bridson and E. R. Smith (1991). When those forms are abbre- viated, as most are, cross references to the corresponding full serial titles are interpolated here alphabetically by abbreviated form. In nomenclatural citations (only), book titles are rendered in the abbreviated forms recommended in F. A. Stafleu and R. S. Cowan (1976–1988) and F. A. Stafleu and E. A. Mennega (1992+). Here, those abbreviated forms are indicated parenthetically following the full citations of the corresponding works, and cross references to the full citations are interpolated in the list alphabetically by abbreviated form. Two or more works published in the same year by the same author or group of coauthors will be distinguished uniquely and consistently throughout all volumes of Flora of North America by lower-case letters (b, c, d, ...) suffixed to the date for the second and subsequent works in the set. The suffixes are assigned in order of editorial encounter and do not reflect chronological sequence of publication. The first work by any particular author or group from any given year carries the implicit date suffix “a”; thus, the sequence of explicit suffixes begins with “b”. Works missing from any suffixed sequence here are ones cited elsewhere in the Flora that are not pertinent in these volumes. -

Natural Heritage Program List of Rare Plant Species of North Carolina 2012
Natural Heritage Program List of Rare Plant Species of North Carolina 2012 Edited by Laura E. Gadd, Botanist John T. Finnegan, Information Systems Manager North Carolina Natural Heritage Program Office of Conservation, Planning, and Community Affairs N.C. Department of Environment and Natural Resources 1601 MSC, Raleigh, NC 27699-1601 Natural Heritage Program List of Rare Plant Species of North Carolina 2012 Edited by Laura E. Gadd, Botanist John T. Finnegan, Information Systems Manager North Carolina Natural Heritage Program Office of Conservation, Planning, and Community Affairs N.C. Department of Environment and Natural Resources 1601 MSC, Raleigh, NC 27699-1601 www.ncnhp.org NATURAL HERITAGE PROGRAM LIST OF THE RARE PLANTS OF NORTH CAROLINA 2012 Edition Edited by Laura E. Gadd, Botanist and John Finnegan, Information Systems Manager North Carolina Natural Heritage Program, Office of Conservation, Planning, and Community Affairs Department of Environment and Natural Resources, 1601 MSC, Raleigh, NC 27699-1601 www.ncnhp.org Table of Contents LIST FORMAT ......................................................................................................................................................................... 3 NORTH CAROLINA RARE PLANT LIST ......................................................................................................................... 10 NORTH CAROLINA PLANT WATCH LIST ..................................................................................................................... 71 Watch Category -

Association Between Chorionicity and Preterm Birth in Twin Pregnancies: a Systematic Review Involving 29 864 Twin Pregnancies
DOI: 10.1111/1471-0528.16479 Systematic Review www.bjog.org Association between chorionicity and preterm birth in twin pregnancies: a systematic review involving 29 864 twin pregnancies S Marleen,a,b C Dias,b R Nandasena,b R MacGregor,c J Allotey,d J Aquilina,c A Khalil,e,f S Thangaratinamg a Barts Research Centre for Women’s Health (BARC), Barts and the London School of Medicine and Dentistry, Queen Mary University of London, London, UK b Sri Jayewardenepura Postgraduate Teaching Hospital, Nugegoda, Sri Lanka c Royal London Hospital, Barts Health NHS Trust, London, UK d Institute of Applied Health Research, University of Birmingham, Birmingham, UK e St George’s University Hospitals NHS Foundation Trust, London, UK f Molecular and Clinical Sciences Research Institute, St George’s Medical School, University of London, London, UK g World Health Organization (WHO) Collaborating Centre for Global Women’s Health, Institute of Metabolism and Systems Research, University of Birmingham, Birmingham, UK Correspondence: S Marleen, Barts Research Centre for Women’s Health (BARC), Barts and the London School of Medicine and Dentistry, Queen Mary University of London, Mile End Road, London E1 4NS, UK. Email: [email protected] Accepted 7 August 2020. Published Online 7 October 2020. Background The perinatal mortality and morbidity among twins I2 = 46%, OR 1.55, 95% CI 1.27–1.89 I2 = 68%, OR 1.47, 95% CI vary by chorionicity. Although it is considered that 1.27–1.69, I2 = 60%, OR 1.66, 95% CI 1.43–1.93, I2 = 65%, monochorionicity is associated with an increased risk of preterm respectively). -

Vascular Plant Inventory of Mount Rainier National Park
National Park Service U.S. Department of the Interior Natural Resource Program Center Vascular Plant Inventory of Mount Rainier National Park Natural Resource Technical Report NPS/NCCN/NRTR—2010/347 ON THE COVER Mount Rainier and meadow courtesy of 2007 Mount Rainier National Park Vegetation Crew Vascular Plant Inventory of Mount Rainier National Park Natural Resource Technical Report NPS/NCCN/NRTR—2010/347 Regina M. Rochefort North Cascades National Park Service Complex 810 State Route 20 Sedro-Woolley, Washington 98284 June 2010 U.S. Department of the Interior National Park Service Natural Resource Program Center Fort Collins, Colorado The National Park Service, Natural Resource Program Center publishes a range of reports that address natural resource topics of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Technical Report Series is used to disseminate results of scientific studies in the physical, biological, and social sciences for both the advancement of science and the achievement of the National Park Service mission. The series provides contributors with a forum for displaying comprehensive data that are often deleted from journals because of page limitations. All manuscripts in the series receive the appropriate level of peer review to ensure that the information is scientifically credible, technically accurate, appropriately written for the intended audience, and designed and published in a professional manner. This report received informal peer review by subject-matter experts who were not directly involved in the collection, analysis, or reporting of the data. -

Pre-Eclampsia/Eclampsia in Twin Pregnancies A
J Med Genet: first published as 10.1136/jmg.13.3.208 on 1 June 1976. Downloaded from Journal of Medical Genetics (1976). 13, 208-211. Pre-eclampsia/eclampsia in twin pregnancies A. McFARLANE and J. S. SCOTT From the Department of Obstetrics and Gynaecology (Leeds Maternity Hospital), University of Leeds, 17 Springfield Mount, Leeds LS2 9NG Summary. A study of 1045 twin gestations with regard to known or likely zygosity and the incidence of pre-eclampsia/eclampsia failed to reveal differences between known dizygous twins and like-sex 'presumed' and 'estimated' monozygous twins except in the 'estimated' data for multigravidae. There was a threefold in- crease in the incidence for twins as opposed to singleton pregnancies. These results are discussed in relation to increased conceptus-mother antigenic differences. It is suggested that the risk of gestosis in twin pregnancy involves more than a summa- tion ofthat operating in two singleton pregnancies. It has been suggested that genetic incompatibility Pregnancies were classified according to definitions given between mother and fetus may be a factor in the below. aetiology of pre-eclampsia (Penrose, 1946; Kalmus, 1946; Platt, Stewart, and Emery, 1958). Epidemio- A. Hypertension status logical evidence has been provided by Stevenson et (1) Mildpre-eclampsia-a basal blood pressure of 120/ 80 mm Hg or less recorded before 24 weeks' gestation, al (1971) in a study of consanguineous marriages in followed by a rise to 140/90 mm Hg or more on at least the Middle East. They also recorded twin data two occasions recorded in the antenatal ward, clinic which pointed to a higher incidence of toxaemia in readings being discounted, together with oedema and unlike-sex as opposed to like-sex twin pregnancies.