Biomedical Applications of Bacteria-Derived Polymers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Bacterial Dissimilation of Citric Acid Carl Robert Brewer Iowa State College
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 1939 Bacterial dissimilation of citric acid Carl Robert Brewer Iowa State College Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Microbiology Commons Recommended Citation Brewer, Carl Robert, "Bacterial dissimilation of citric acid " (1939). Retrospective Theses and Dissertations. 13227. https://lib.dr.iastate.edu/rtd/13227 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. INFORMATION TO USERS This manuscript has been reproduced from the microfilm master. UMI films the text directly from the original or copy submitted. Thus, some thesis and dissertation copies are in typewriter face, while others may be from any type of computer printer. The quality of this reproduction is dependent upon the quality of the copy submitted. Brol<en or indistinct print, colored or poor quality illustrations and photographs, print bleedthrough, substandard margins, and improper alignment can adversely affect reproduction. In the unlikely event that the author did not send UMI a complete manuscript and there are missing pages, these will be noted. Also, if unauthorized copyright material had to be removed, a note will indicate the deletion. Oversize materials (e.g., maps, drawings, charts) are reproduced by sectioning the original, beginning at the upper left-hand comer and continuing from left to right in equal sections with small overlaps. -

Congenital Disorders of Glycosylation from a Neurological Perspective
brain sciences Review Congenital Disorders of Glycosylation from a Neurological Perspective Justyna Paprocka 1,* , Aleksandra Jezela-Stanek 2 , Anna Tylki-Szyma´nska 3 and Stephanie Grunewald 4 1 Department of Pediatric Neurology, Faculty of Medical Science in Katowice, Medical University of Silesia, 40-752 Katowice, Poland 2 Department of Genetics and Clinical Immunology, National Institute of Tuberculosis and Lung Diseases, 01-138 Warsaw, Poland; [email protected] 3 Department of Pediatrics, Nutrition and Metabolic Diseases, The Children’s Memorial Health Institute, W 04-730 Warsaw, Poland; [email protected] 4 NIHR Biomedical Research Center (BRC), Metabolic Unit, Great Ormond Street Hospital and Institute of Child Health, University College London, London SE1 9RT, UK; [email protected] * Correspondence: [email protected]; Tel.: +48-606-415-888 Abstract: Most plasma proteins, cell membrane proteins and other proteins are glycoproteins with sugar chains attached to the polypeptide-glycans. Glycosylation is the main element of the post- translational transformation of most human proteins. Since glycosylation processes are necessary for many different biological processes, patients present a diverse spectrum of phenotypes and severity of symptoms. The most frequently observed neurological symptoms in congenital disorders of glycosylation (CDG) are: epilepsy, intellectual disability, myopathies, neuropathies and stroke-like episodes. Epilepsy is seen in many CDG subtypes and particularly present in the case of mutations -

Citrobacter Koseri, Levinea Malonatica
INTERNATIONALJOURNAL OF SYSTEMATICBACTERIOLOGY, Jan. 1990, p. 107-108 Vol. 40, No. 1 0020-7713/90/010107-02$02.oo/o Copyright 0 1990, International Union of Microbiological Societies Correct Names of the Species Citrobacter koseri, Levinea malonatica , and Citrobacter diversus Request for an Opinion WILHELM FREDERIKSEN Department of Diagnostic Bacteriology and Antibiotics, Statens Seruminstitut, DK-2300 Copenhagen S,Denmark The single species carrying the three names Citrobacter koseri, Levinea malonatica, and Citrobacter diversus differs by at least eight characteristics from Citrobacter diversum as described by Werkman and Gillen in 1932. It is obviously not the same organism. Accordingly, the species should not carry the name proposed by Werkman and Gillen. I request that the name Citrobacter diversus be placed on the list of nomina rejicienda. Citrobacter koseri is the correct name. Levinea koseri is a correct combination when the genus Levinea is accepted. The epithet maZoonatica is a later synonym of the epithet koseri. In 1932 Werkman and Gillen (7) proposed a new genus, taxon C. diversum described by Werkman and Gillen in the Citrobacter, containing seven species. The description of following characteristics: motility, production of H,S, and Citrobacter diversum (sic) of Werkman and Gillen was based production of acid from inositol and raffinose. To this can be on two strains. These strains have not been kept available in added production of acid from glycogen, melizitose, starch, collections, and the name C. diversum did not come into and galactose, as Table 1 shows. general use. Ewing and Davis obviously accepted four deviations to In 1970 Frederiksen (4) described a new species which he lead to the statement about reactions “similar to those named Citrobacter koseri. -

Viruses Like Sugars: How to Assess Glycan Involvement in Viral Attachment
microorganisms Review Viruses Like Sugars: How to Assess Glycan Involvement in Viral Attachment Gregory Mathez and Valeria Cagno * Institute of Microbiology, Lausanne University Hospital, University of Lausanne, 1011 Lausanne, Switzerland; [email protected] * Correspondence: [email protected] Abstract: The first step of viral infection requires interaction with the host cell. Before finding the specific receptor that triggers entry, the majority of viruses interact with the glycocalyx. Identifying the carbohydrates that are specifically recognized by different viruses is important both for assessing the cellular tropism and for identifying new antiviral targets. Advances in the tools available for studying glycan–protein interactions have made it possible to identify them more rapidly; however, it is important to recognize the limitations of these methods in order to draw relevant conclusions. Here, we review different techniques: genetic screening, glycan arrays, enzymatic and pharmacological approaches, and surface plasmon resonance. We then detail the glycan interactions of enterovirus D68 and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), highlighting the aspects that need further clarification. Keywords: attachment receptor; viruses; glycan; sialic acid; heparan sulfate; HBGA; SARS-CoV-2; EV-D68 Citation: Mathez, G.; Cagno, V. Viruses Like Sugars: How to Assess 1. Introduction Glycan Involvement in Viral This review focuses on methods for assessing the involvement of carbohydrates in Attachment. Microorganisms 2021, 9, viral attachment and entry into the host cell. Viruses often bind to entry receptors that are 1238. https://doi.org/10.3390/ not abundant on the cell surface; to increase their chances of finding them, they initially microorganisms9061238 bind to attachment receptors comprising carbohydrates that are more widely expressed. -

Prenatal Testing Requisition Form
BAYLOR MIRACA GENETICS LABORATORIES SHIP TO: Baylor Miraca Genetics Laboratories 2450 Holcombe, Grand Blvd. -Receiving Dock PHONE: 800-411-GENE | FAX: 713-798-2787 | www.bmgl.com Houston, TX 77021-2024 Phone: 713-798-6555 PRENATAL COMPREHENSIVE REQUISITION FORM PATIENT INFORMATION NAME (LAST,FIRST, MI): DATE OF BIRTH (MM/DD/YY): HOSPITAL#: ACCESSION#: REPORTING INFORMATION ADDITIONAL PROFESSIONAL REPORT RECIPIENTS PHYSICIAN: NAME: INSTITUTION: PHONE: FAX: PHONE: FAX: NAME: EMAIL (INTERNATIONAL CLIENT REQUIREMENT): PHONE: FAX: SAMPLE INFORMATION CLINICAL INDICATION FETAL SPECIMEN TYPE Pregnancy at risk for specific genetic disorder DATE OF COLLECTION: (Complete FAMILIAL MUTATION information below) Amniotic Fluid: cc AMA PERFORMING PHYSICIAN: CVS: mg TA TC Abnormal Maternal Screen: Fetal Blood: cc GESTATIONAL AGE (GA) Calculation for AF-AFP* NTD TRI 21 TRI 18 Other: SELECT ONLY ONE: Abnormal NIPT (attach report): POC/Fetal Tissue, Type: TRI 21 TRI 13 TRI 18 Other: Cultured Amniocytes U/S DATE (MM/DD/YY): Abnormal U/S (SPECIFY): Cultured CVS GA ON U/S DATE: WKS DAYS PARENTAL BLOODS - REQUIRED FOR CMA -OR- Maternal Blood Date of Collection: Multiple Pregnancy Losses LMP DATE (MM/DD/YY): Parental Concern Paternal Blood Date of Collection: Other Indication (DETAIL AND ATTACH REPORT): *Important: U/S dating will be used if no selection is made. Name: Note: Results will differ depending on method checked. Last Name First Name U/S dating increases overall screening performance. Date of Birth: KNOWN FAMILIAL MUTATION/DISORDER SPECIFIC PRENATAL TESTING Notice: Prior to ordering testing for any of the disorders listed, you must call the lab and discuss the clinical history and sample requirements with a genetic counselor. -

Carbapenem-Resistant Enterobacteriaceae a Microbiological Overview of (CRE) Carbapenem-Resistant Enterobacteriaceae
PREVENTION IN ACTION MY bugaboo Carbapenem-resistant Enterobacteriaceae A microbiological overview of (CRE) carbapenem-resistant Enterobacteriaceae. by Irena KennelEy, PhD, aPRN-BC, CIC This agar culture plate grew colonies of Enterobacter cloacae that were both characteristically rough and smooth in appearance. PHOTO COURTESY of CDC. GREETINGS, FELLOW INFECTION PREVENTIONISTS! THE SCIENCE OF infectious diseases involves hundreds of bac- (the “bug parade”). Too much information makes it difficult to teria, viruses, fungi, and protozoa. The amount of information tease out what is important and directly applicable to practice. available about microbial organisms poses a special problem This quarter’s My Bugaboo column will feature details on the CRE to infection preventionists. Obviously, the impact of microbial family of bacteria. The intention is to convey succinct information disease cannot be overstated. Traditionally the teaching of to busy infection preventionists for common etiologic agents of microbiology has been based mostly on memorization of facts healthcare-associated infections. 30 | SUMMER 2013 | Prevention MULTIDRUG-resistant GRAM-NEGative ROD ALert: After initial outbreaks in the northeastern U.S., CRE bacteria have THE CDC SAYS WE MUST ACT NOW! emerged in multiple species of Gram-negative rods worldwide. They Carbapenem-resistant Enterobacteriaceae (CRE) infections come have created significant clinical challenges for clinicians because they from bacteria normally found in a healthy person’s digestive tract. are not consistently identified by routine screening methods and are CRE bacteria have been associated with the use of medical devices highly drug-resistant, resulting in delays in effective treatment and a such as: intravenous catheters, ventilators, urinary catheters, and high rate of clinical failures. -

F04b57ca351b0dc3389ebe992c
Review Insights into complexity of congenital disorders of glycosylation Sandra Supraha Goreta*, Sanja Dabelic, Jerka Dumic University of Zagreb, Faculty of Pharmacy and Biochemistry, Department of Biochemistry and Molecular Biology, Zagreb, Croatia *Corresponding author: [email protected] Abstract Biochemical and biological properties of glycoconjugates are strongly determined by the specifi c structure of its glycan parts. Glycosylation, the covalent attachment of sugars to proteins and lipids, is very complex and highly-coordinated process involving > 250 gene products. Defi ciency of glycosylation enzymes or transporters results in impaired glycosylation, and consequently pathological modulation of many physiological processes. Inborn defects of glycosylation enzymes, caused by the specifi cmutations, lead to the development of rare, but severe diseases – congenital disor- ders of glycosylation (CDGs). Up today, there are more than 45 known CDGs. Their clinical manifestations range from very mild to extremely severe (even lethal) and unfortunately, only three of them can be eff ectively treated nowadays. CDG symptoms highly vary, though someare common for several CDG types but also for other unrelated diseases, especially neurological ones, leaving the possibility that many CDGs cases are under- or mis- diagnosed. Glycan analysis of serum transferrin (by isoelectric focusing or more sophisticated methods, such as HPLC (high-performance liquid chro- matography) or MALDI (matrix-assisted laser desorption/ionization)) or serum N-glycans -

Metabolic Enzyme Expression Highlights a Key Role for MTHFD2 and the Mitochondrial Folate Pathway in Cancer
ARTICLE Received 1 Nov 2013 | Accepted 17 Dec 2013 | Published 23 Jan 2014 DOI: 10.1038/ncomms4128 Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer Roland Nilsson1,2,*, Mohit Jain3,4,5,6,*,w, Nikhil Madhusudhan3,4,5, Nina Gustafsson Sheppard1,2, Laura Strittmatter3,4,5, Caroline Kampf7, Jenny Huang8, Anna Asplund7 & Vamsi K. Mootha3,4,5 Metabolic remodeling is now widely regarded as a hallmark of cancer, but it is not clear whether individual metabolic strategies are frequently exploited by many tumours. Here we compare messenger RNA profiles of 1,454 metabolic enzymes across 1,981 tumours spanning 19 cancer types to identify enzymes that are consistently differentially expressed. Our meta- analysis recovers established targets of some of the most widely used chemotherapeutics, including dihydrofolate reductase, thymidylate synthase and ribonucleotide reductase, while also spotlighting new enzymes, such as the mitochondrial proline biosynthetic enzyme PYCR1. The highest scoring pathway is mitochondrial one-carbon metabolism and is centred on MTHFD2. MTHFD2 RNA and protein are markedly elevated in many cancers and correlated with poor survival in breast cancer. MTHFD2 is expressed in the developing embryo, but is absent in most healthy adult tissues, even those that are proliferating. Our study highlights the importance of mitochondrial compartmentalization of one-carbon metabolism in cancer and raises important therapeutic hypotheses. 1 Unit of Computational Medicine, Department of Medicine, Karolinska Institutet, 17176 Stockholm, Sweden. 2 Center for Molecular Medicine, Karolinska Institutet, 17176 Stockholm, Sweden. 3 Broad Institute, Cambridge, Massachusetts 02142, USA. 4 Department of Systems Biology, Harvard Medical School, Boston, Massachusetts 02115, USA. -

Genome-Wide Investigation of Cellular Functions for Trna Nucleus
Genome-wide Investigation of Cellular Functions for tRNA Nucleus- Cytoplasm Trafficking in the Yeast Saccharomyces cerevisiae DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Hui-Yi Chu Graduate Program in Molecular, Cellular and Developmental Biology The Ohio State University 2012 Dissertation Committee: Anita K. Hopper, Advisor Stephen Osmani Kurt Fredrick Jane Jackman Copyright by Hui-Yi Chu 2012 Abstract In eukaryotic cells tRNAs are transcribed in the nucleus and exported to the cytoplasm for their essential role in protein synthesis. This export event was thought to be unidirectional. Surprisingly, several lines of evidence showed that mature cytoplasmic tRNAs shuttle between nucleus and cytoplasm and their distribution is nutrient-dependent. This newly discovered tRNA retrograde process is conserved from yeast to vertebrates. Although how exactly the tRNA nuclear-cytoplasmic trafficking is regulated is still under investigation, previous studies identified several transporters involved in tRNA subcellular dynamics. At least three members of the β-importin family function in tRNA nuclear-cytoplasmic intracellular movement: (1) Los1 functions in both the tRNA primary export and re-export processes; (2) Mtr10, directly or indirectly, is responsible for the constitutive retrograde import of cytoplasmic tRNA to the nucleus; (3) Msn5 functions solely in the re-export process. In this thesis I focus on the physiological role(s) of the tRNA nuclear retrograde pathway. One possibility is that nuclear accumulation of cytoplasmic tRNA serves to modulate translation of particular transcripts. To test this hypothesis, I compared expression profiles from non-translating mRNAs and polyribosome-bound translating mRNAs collected from msn5Δ and mtr10Δ mutants and wild-type cells, in fed or acute amino acid starvation conditions. -

CDG-Ih) (ALG8 Deficiency
550 LETTER TO JMG J Med Genet: first published as 10.1136/jmg.2003.016923 on 2 July 2004. Downloaded from Clinical and molecular features of three patients with congenital disorders of glycosylation type Ih (CDG-Ih) (ALG8 deficiency) E Schollen, C G Frank, L Keldermans, R Reyntjens, C E Grubenmann, P T Clayton, B G Winchester, J Smeitink, R A Wevers, M Aebi, T Hennet, G Matthijs ............................................................................................................................... J Med Genet 2004;41:550–556. doi: 10.1136/jmg.2003.016923 rotein glycosylation is an essential post-translational Key points modification of various proteins, affecting their folding, Psorting, and function. Inborn defects in the assembly and processing of glycans on glycoproteins are known as N Congenital disorder of glycosylation type Ih (CDG-Ih) congenital disorders of glycosylation (CDG) and can affect is caused by a defect in the dolichyl-P-Glc:Glc1 both N- and O-glycosylation (for reviews see Marquardt and Man9GlcNAc2-PP-dolichyl a1,3-glucosyltransferase Denecke,1 Jaeken,2 and Grunewald et al3). N-glycosylation (ALG8). Thus far, the only patient has been described defects and especially defects in the assembly of the dolichol with a relatively mild clinical picture. linked N-glycan precursor in the endoplasmic reticulum (ER) N We describe the molecular and clinical features of (CDG-I) result in hypoglycosylation of many kinds of three patients from two families with an ALG8 (serum) glycoproteins. CDG-I is therefore a group of multi- deficiency. All three patients presented with severe, systemic disorders. life-threatening multi organ failure and died within The assembly of the N-glycan precursor in the ER is a their first months of life. -
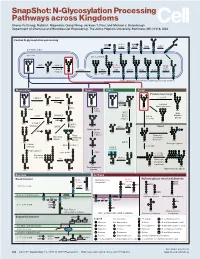
N-Glycosylation Processing Pathways Across Kingdoms Cheng-Yu Chung, Natalia I
SnapShot: N-Glycosylation Processing Pathways across Kingdoms Cheng-Yu Chung, Natalia I. Majewska, Qiong Wang, Jackson T. Paul, and Michael J. Betenbaugh Department of Chemical and Biomolecular Engineering, The Johns Hopkins University, Baltimore, MD 21218, USA Central N-glycosylation processing UDP ALG13 GDP ALG7 ALG14 ALG1 ALG2 ALG11 P P P P P CYTOPLASM P P P P P P GOLGI ER LUMEN GNTI α -Man IA En Bloc ALG3 Transfer ALG9 α-Man IB ER Man I α -Glc I OST ALG10 ALG8 ALG6 ALG12 Asn α -Man IC Asn Asn Asn α -Glc II P P P P P Asn P P P P P ALGs: asparagine-linked N-glycosylation processing enzymes Mammalian Insect Plant Fungi Filamentous fungi α-Man II FUT8 α-1,2-MT XYLT GDP Terminal Asn Asn Asn Asn UDP Asn Galactofuranose GNTII Asn Asn FUT8 f ± α-Gal Asn FUTC3 GNTII High FUT11 α -GALT B14GALTI Och1p Mannose FUT12 glycan α-Man II α-1,2-MT UDP Asn Asn ST3GAL4 Asn Asn GNTIV ST6GAL1 GNTIII GNTV Asn GNase Yeast Hybrid glycan P CMP GNase Asn Asn Tetra- α-Man II Mannosyl- Bisecting antennary phosphate glycan glycan transferase Asn Asn Asn Asn GALT1 Asn ST8SIA2 Core type glycan GNTs B14GALTI α-1,6-MT ST8SIA4 Asn Lewis A structure Polysialylation α-Man PolyLacNAc α-1,2-MT Alternative α-1,3-MT P sialic acid FUT13 ] [ STs Mannosyl- n phosphate Asn Asn Asn transferase Asn Asn Paucimannose Hypermannose glycan Asn Asn Asn glycans Bacteria Archaea Archaea glycan structural diversity Block transfer En Bloc Methanococcus En Bloc Transfer Transfer maripaludis OCH3 Thr OCH AgIB Thr OCH3 3 PERIPLASM PglB B B Asn P - P Asn OSO3 P Asn Asn INNER MEMBRANE P Asn P -

Citrobacter Koseri. II. Serological and Biochemical Examina- Tion of Citrobacter Koseri Strains from Clinical Specimens by B
J. Hyg., Camb. (1975), 75, 129 129 Printed in Great Britain Citrobacter koseri. II. Serological and biochemical examina- tion of Citrobacter koseri strains from clinical specimens BY B. ROWE, R. J. GROSS AND H. A. ALLEN Salmonella and Shigella Reference Laboratory, Central Public Health Laboratory, Colindale Avenue, London NW9 5HT (Received 30 January 1975) SUMMLARY 165 strains of Citrobacter koseri isolated from clinical specimens were studied and their biochemical reactions determined. They were examined serologically by means of a scheme consisting of 14 0 antigens. The sources of the clinical specimens were tabulated and the epidemiological information was summarized. The clinical significance of these findings is discussed. INTRODUCTION Bacterial strains variously described as Citrobacter koseri (Frederiksen, 1970), Citrobacter diversus (Ewing & Davis, 1972) and Levinea malonatica (Young, Kenton, Hobbs & Moody, 1971) should be regarded as members of a single species (Gross & Rowe, 1974). In this publication the name C. koseri has been used for convenience only, pending an agreement on the nomenclature of these organisms. In most genera within the family Enterobacteriaceae, serotyping has been invaluable for the precise identification of strains required in epidemiological investigations. An antigenic scheme for C. koseri has been described (Gross & Rowe, 1974) which initially consisted of seven 0 antigens and was subsequently expanded to 14 0 antigens (Gross & Rowe, 1975). The present study involves the examination of 165 strains from clinical sources. Their biochemical reactions and somatic 0 antigens have been determined and the clinical and epidemiological significance of these findings is discussed. MATERIALS AND METHODS Strains The sources of 165 strains isolated from clinical specimens are shown in Table 1.