Parthenocarpy and Functional Sterility in Tomato
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

New Acid Citrus Selections for Florida
New acid citrus selections for Florida Lemon and lime-like selections with niche market potential are being developed with biotechnology at the University of Florida. By Jude Grosser, Zenaida Viloria and Manjul Dutt re you ready for a purple margarita? Would you like a fragrant, juicy lime is a naturally occurring citrus lemon for your iced tea with no seeds to clog your straw or dishwasher triploid, which is why it is seedless. drain? How about some seedless lime trees that are cold-hardy enough The new seedless watermelons in the Afor Central Florida? These and many more interesting acid-citrus marketplace are also triploids. selections are now on the horizon, including some with good ornamental potential. Due to the fact that new and This article will focus on progress in using emerging biotechnologies to develop improved citrus cultivars must be interesting new citrus cultivars in the lemon and lime group. Cultivars include seedless to compete in the national some that will not have regulatory constraints, and also a genetically modified and international marketplace, the organism (GMO)-derived purple Key lime as a teaser for the future. University of Florida’s Citrus Research and Education Center (UF/CREC) LEARNING FROM they are triploids. People and most citrus improvement team (working THE BANANA citrus trees are diploid, meaning with Fred Gmitter) has formulated Have you ever wondered why you there are two sets of chromosomes in several ways to create triploids as a key never find seeds in your bananas? Did each cell. Triploid bananas have three method of developing seedless citrus you know that there are wild-type sets of chromosomes per cell. -

The Satsuma Mandarin1 Peter C
HS195 The Satsuma Mandarin1 Peter C. Andersen and James J. Ferguson2 Scientific Name Family Citrus unshiu Marcovitch Rutaceae Common Name Origin In most citrus producing areas, satsuma mandarin is the China and Japan preferred name, but satsuma tangerine is also used (Figures 1 and 2). Distribution Satsumas are grown in cool subtropical regions of Japan, Spain, central China, Korea, Turkey, along the Black Sea in Russia, southern South Africa, South America, and on a small scale in central California and northern Florida. The world’s largest satsuma industry is located in southern Ja- pan where climatic conditions are favorable for the produc- tion of early ripening satsuma mandarins of high quality. Selection of slight mutations and seedlings from controlled pollinations over many years has resulted in a collection of Figure 1. A satsuma orchard in north Florida. over 100 cultivars that differ in date of maturity, fruit shape, Credits: P. C. Andersen color, and quality. ‘Owari’ is the primary satsuma cultivar commercially grown in Florida, but ‘Brown Select’, ‘Early St. Ann’, ‘Silverhill’, and ‘Kimbrough’ are also available. History Satsuma mandarin may have originated in China, but it was first reported in Japan more than 700 years ago, where it is now the major citrus species grown. The first recorded introduction into the United States was in Florida by George R. Hall in 1876. The name “satsuma” is credited to Figure 2. Mature satsumas ready for harvest. the wife of a United States minister to Japan, General Van Credits: P. C. Andersen 1. This document is HS195, one of a series of the Horticultural Sciences Department, UF/IFAS Extension. -

UC Riverside Electronic Theses and Dissertations
UC Riverside UC Riverside Electronic Theses and Dissertations Title A Molecular Genetic Approach to Evaluate a Novel Seedless Phenotype Found in Tango, a New Variety of Mandarin Developed From Gamma-Irradiated W. Murcott Permalink https://escholarship.org/uc/item/1gp2m8vb Author Crowley, Jennifer Robyn Publication Date 2011 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA RIVERSIDE A Molecular Genetic Approach to Evaluate a Novel Seedless Phenotype Found in Tango, a New Variety of Mandarin Developed From Gamma-Irradiated W. Murcott Doctor of Philosophy in Genetics, Genomics and Bioinformatics by Jennifer Robyn Crowley December 2011 Dissertation Committee: Dr. Mikeal Roose, Chairperson Dr. Linda Walling Dr. Katherine Borkovich Copyright by Jennifer Robyn Crowley 2011 The Dissertation of Jennifer Robyn Crowley is approved: ________________________________________________________________________ ________________________________________________________________________ Committee Chairperson University of California, Riverside Acknowledgments I would like to acknowledge and give my many thanks to several people who contributed both their time and their effort towards my dissertation work. I would like to thank Dr. David Carter for his instruction regarding the correct use of the Leica SP2 confocal microscope, as well as for his help in gathering quality computerized images. I would like to thank Dr. Chandrika Ramadugu and Dr. Manjunath Keremane for their assistance with the use of the Stratagene MX thermocycler, as well as for their help in qPCR data interpretation. To Dr. Chandrika Ramadugu, I would also like to extend my gratitude for the contribution she made towards my research project by providing me with clones of a DNA sequence of interest. -

Improvement of Subtropical Fruit Crops: Citrus
IMPROVEMENT OF SUBTROPICAL FRUIT CROPS: CITRUS HAMILTON P. ÏRAUB, Senior Iloriiciilturist T. RALPH ROBCNSON, Senior Physiolo- gist Division of Frnil and Vegetable Crops and Diseases, Bureau of Plant Tndusiry MORE than half of the 13 fruit crops known to have been cultivated longer than 4,000 years,according to the researches of DeCandolle (7)\ are tropical and subtropical fruits—mango, oliv^e, fig, date, banana, jujube, and pomegranate. The citrus fruits as a group, the lychee, and the persimmon have been cultivated for thousands of years in the Orient; the avocado and papaya were important food crops in the American Tropics and subtropics long before the discovery of the New World. Other types, such as the pineapple, granadilla, cherimoya, jaboticaba, etc., are of more recent introduction, and some of these have not received the attention of the plant breeder to any appreciable extent. Through the centuries preceding recorded history and up to recent times, progress in the improvement of most subtropical fruits was accomplished by the trial-error method, which is crude and usually expensive if measured by modern standards. With the general accept- ance of the Mendelian principles of heredity—unit characters, domi- nance, and segregation—early in the twentieth century a starting point was provided for the development of a truly modern science of genetics. In this article it is the purpose to consider how subtropical citrus fruit crops have been improved, are now being improved, or are likel3^ to be improved by scientific breeding. Each of the more important crops will be considered more or less in detail. -

Parthenocarpy in Summer Squash on a Lima Silt Loam (Fine-Loamy, Mixed, Mesic, Glossoboric Hapludalf) at Geneva, N.Y
HORTSCIENCE 34(4):715–717. 1999. Materials and Methods Experiments were conducted for 4 years Parthenocarpy in Summer Squash on a Lima silt loam (fine-loamy, mixed, mesic, Glossoboric Hapludalf) at Geneva, N.Y. Four- 1 2 R.W. Robinson and Stephen Reiners week-old transplants were set in the field and Department of Horticultural Sciences, New York State Agricultural Experiment spaced at 0.9 m in rows on 1.5-m centers. Station, Geneva, NY 14456-0462 Weeds were controlled using recommended herbicides, cultivation, and hand weeding, Additional index words. Cucurbita pepo, fruit set, seedless fruit, row covers, pollination while insect and disease pressure was moni- tored and protective treatments applied when Abstract. Summer squash (Cucurbita pepo L.) cultivars were compared for ability to set warranted (Becker, 1992). parthenocarpic fruit. Some cultivars set no parthenocarpic fruit and others varied in the Over 95% of the first pistillate flowers to amount of fruit set when not pollinated. The degree of parthenocarpy varied with season, develop were closed before anthesis by plac- but the relative ranking of cultivars for parthenocarpy was generally similar. Cultivars ing plastic-covered wire around the unopened with the best parthenocarpic fruit set were of the dark green, zucchini type, but some petals to prevent insect pollination. Any open cultivars of other fruit types also set parthenocarpic fruit. A summer squash cultivar was flowers were removed within 2 d of anthesis to developed that combines a high rate of natural parthenocarpy with multiple disease prevent development of open-pollinated fruit. resistance. Yield of summer squash plants grown under row covers that excluded Fruit from closed flowers were harvested im- pollinating insects was as much as 83% of that of insect-pollinated plants in the open. -

New and Noteworthy Citrus Varieties Presentation
New and Noteworthy Citrus Varieties Citrus species & Citrus Relatives Hundreds of varieties available. CITRON Citrus medica • The citron is believed to be one of the original kinds of citrus. • Trees are small and shrubby with an open growth habit. The new growth and flowers are flushed with purple and the trees are sensitive to frost. • Ethrog or Etrog citron is a variety of citron commonly used in the Jewish Feast of Tabernacles. The flesh is pale yellow and acidic, but not very juicy. The fruits hold well on the tree. The aromatic fruit is considerably larger than a lemon. • The yellow rind is glossy, thick and bumpy. Citron rind is traditionally candied for use in holiday fruitcake. Ethrog or Etrog citron CITRON Citrus medica • Buddha’s Hand or Fingered citron is a unique citrus grown mainly as a curiosity. The six to twelve inch fruits are apically split into a varying number of segments that are reminiscent of a human hand. • The rind is yellow and highly fragrant at maturity. The interior of the fruit is solid rind with no flesh or seeds. • Fingered citron fruits usually mature in late fall to early winter and hold moderately well on the tree, but not as well as other citron varieties. Buddha’s Hand or Fingered citron NAVEL ORANGES Citrus sinensis • ‘Washington navel orange’ is also known • ‘Lane Late Navel’ was the first of a as the Bahia. It was imported into the number of late maturing Australian United States in 1870. navel orange bud sport selections of Washington navel imported into • These exceptionally delicious, seedless, California. -

Review Paper POLYPLOIDY in VEGETABLES Journal of Global
Journal of Global Biosciences ISSN 2320-1355 Volume 4, Number 3, 2015, pp. 1774-1779 Website: www.mutagens.co.in E-mail: [email protected] [email protected] Review Paper POLYPLOIDY IN VEGETABLES N.A. Kazi Department of Horticulture, College of Agriculture, Pune 411 005 India . Abstract Polyploids are organisms with three or more complete chromosome sets. Polyploids were marked with phenotypic and genotypic variations. Beneficial variations can be further used in plant breeding programmes. Several abnormalities were also reported. Successful induction of polyploidy can also result in change in taste. In watermelons sweetness order was 3x > 4x > 2x. Polyploidy was found to be induced by colchicine, colchamine, oryzalin, colcemid, trifluralin etc. Chromosome doubling was achieved by different concentrations of colchicine. Review of polyploidy in vegetable crops Viz., Cabbage, Brussels sprouts, Pea, Watermelon, Muskmelon, Onion, Garlic, Carrot, Radish, Cocoyam, Pointed gourd, Bitter Gourd, Snake gourd, Fluted pumpkin, Cucumber, And French Bean was undertaken. Key words: Polyploidy, Vegetables, Breeding, Colchicine. INTRODUCTION Polyploids are organisms with three or more complete chromosome sets. Polyploidization is widespread in plants and animals, and is an important mechanism of speciation. The formation of fertile polyploids not only promoted the interflow of genetic materials among species and enriched the species diversity, but also laid the foundation for polyploidy breeding. The study of polyploids has both important theoretical significance and valuable applications. The production and application of polyploidy breeding have brought remarkable economic and social benefits. (Can, 2012). Polyploidy is responsible for the creation of thousands of species in today's planet, and will continue to do so. -

The Origins of Fruits, Fruit Growing, and Fruit Breeding
The Origins of Fruits, Fruit Growing, and Fruit Breeding Jules Janick Department of Horticulture and Landscape Architecture Purdue University 625 Agriculture Mall Drive West Lafayette, Indiana 47907-2010 I. INTRODUCTION A. The Origins of Agriculture B. Origins of Fruit Culture in the Fertile Crescent II. THE HORTICULTURAL ARTS A. Species Selection B. Vegetative Propagation C. Pollination and Fruit Set D. Irrigation E. Pruning and Training F. Processing and Storage III. ORIGIN, DOMESTICATION, AND EARLY CULTURE OF FRUIT CROPS A. Mediterranean Fruits 1. Date Palm 2. Olive 3. Grape 4. Fig 5. Sycomore Fig 6. Pomegranate B. Central Asian Fruits 1. Pome Fruits 2. Stone fruits C. Chinese and Southeastern Asian Fruits 1. Peach 1 2. Citrus 3. Banana and Plantain 4. Mango 5. Persimmon 6. Kiwifruit D. American Fruits 1. Strawberry 2. Brambles 3. Vacciniums 4. Pineapple 5. Avocado 6. Papaya IV. GENETIC CHANGES AND CULTURAL FACTORS IN DOMESTICATION A. Mutations as an Agent of Domestication B. Interspecific Hybridization and Polyploidization C. Hybridization and Selection D. Champions E. Lost Fruits F. Fruit Breeding G. Predicting Future Changes I. INTRODUCTION Crop plants are our greatest heritage from prehistory (Harlan 1992; Diamond 2002). How, where, and when the domestication of crops plants occurred is slowly becoming revealed although not completely understood (Camp et al. 1957; Smartt and Simmonds 1995; Gepts 2003). In some cases, the genetic distance between wild and domestic plants is so great, maize and crucifers, for example, that their origins are obscure. The origins of the ancient grains (wheat, maize, rice, and sorghum) and pulses (sesame and lentil) domesticated in Neolithic times have been the subject of intense interest and the puzzle is being solved with the new evidence based on molecular biology (Gepts 2003). -

Seedless Orange: a New Mutant with Female Sterility
371 Scientia Agricola http://dx.doi.org/10.1590/1678-992X-2016-0129 ‘URS Campestre’ seedless orange: a new mutant with female sterility Renar João Bender1*, Rinaldo Pires dos Santos2, Divanilde Guerra3, Sergio Francisco Schwarz1, Stefan da Silveira Bender4 1Federal University of Rio Grande do Sul − Horticultural ABSTRACT: Seedlessness in fruit is a trait that is much sought after by juice making industries. Sciences Dept., Av. Bento Gonçalves, 7712 − 91540-000 − Close to the city of São Sebastião do Caí, in the state of Rio Grande do Sul (RS), Brazil, a new Porto Alegre, RS − Brazil. mutant orange originating from natural mutation was identified and selected as a seedless 2Federal University of Rio Grande do Sul − Biosciences material. To determine the mechanisms involved in the absence of seeds, the reproductive Institute − Dept. of Botany. Av. Bento Gonçalves, 9500 - structures of this new mutant by comparison with a Valencia sweet orange as control, a culti- 91501-970 − Porto Alegre, RS − Brazil. var with a profusion of seeds, was analyzed in terms of meiotic behavior, meiotic index, pollen 3Rio Grande do Sul State University – Campus Três Passos, viability, in vitro germination, and ovule features to determine the grounds for seed absence. R. Cipriano Barata 47, Três Passos – 98600-000, RS – Other morphological analyzes allowed for visualizing the structures of normal appearance Brazil. and size in both cultivars. Meiotic analysis identified chromosome normal pairing with a pre- 4Federal University of Rio Grande do Sul/Faculty of dominance of bivalents at diakinesis and metaphase 1. URS Campestre flowers at different Agronomy. developmental stages had anthers and ovaries whose dimensions are typical while pollen grain *Corresponding author <[email protected]> analysis pointed to a standard developmental pattern, normal meiosis, high viability (84 %) and elevated in vitro pollen tube germination rates (63 %). -

No Effect of Seed Presence Or Absence on Sugar Content And
HORTSCIENCE 53(3):304–312. 2018. https://doi.org/10.21273/HORTSCI12707-17 Nakai]: the seeded diploid ‘Hitorijime- BonBon’ and the seedless triploid ‘Sandia’ (Hagihara Farm Co., Ltd., Nara, Japan). Both No Effect of Seed Presence or Absence cultivars are suitable for harvesting by 40 d after pollination. We obtained seedlings in on Sugar Content and Water Status of May 2015–17 and transplanted them into three hydroponic culture beds (258 cm · Seeded and Seedless Watermelon Fruits 110 cm · 15 cm) at 50 cm between plants (5 plants · 2 lines were grown in each bed). Sachiko Kawamura and Kyoko Ida Nutrient solution (Otsuka Chemical Co., School of Agriculture, Meiji University, Kawasaki 214-8571, Japan Ltd., Osaka, Japan) with an electric conduc- tivity (EC) of 1.2 mS·cm–1 and a pH of 5.8 Masako Osawa was maintained at a depth of 10 cm using EC Hagihara Farm Co., Ltd., Tawaramoto, Nara 636-0222, Japan meter (Atago Co., Ltd., Tokyo, Japan) and pH meter (Thermo Fisher Scientific K.K., Takashi Ikeda1 Kanagawa, Japan). The beds were maintained School of Agriculture, Meiji University, Kawasaki 214-8571, Japan in a glasshouse that was ventilated whenever the air temperature exceeded 25 °C. Additional index words. HPLC, isopiestic psychrometer, osmotic potential, water potential In 2015 (Expt. 1), female flowers of each Abstract. We investigated physiological differences in watermelon [Citrullus lanatus cultivar were pollinated conventionally by (Thunb.) Matsum. et Nakai] fruits among seeded diploid and seedless triploid fruits, hand. In 2016 (Expt. 2), half of the female N-(2-chloro-4-pyridyl)-N#-phenylurea (CPPU)–treated seedless fruits, and soft-X– flowers of ‘Hitorijime-BonBon’ were pol- irradiated pollen-pollinated seedless fruits to investigate the effect of the presence or linated conventionally and the other half were sprayed with 0.5 mL of CPPU solution absence of seeds on water relations and sugar content. -

Growing Seedless Watermelon 1
Archival copy: for current recommendations see http://edis.ifas.ufl.edu or your local extension office. HS687 Growing Seedless Watermelon 1 Donald N. Maynard2 Fruit of standard seeded watermelon varieties may contain as many as 1,000 seeds in each fruit ( Plate 1 ). The presence of seeds throughout the flesh makes the removal of seeds while eating difficult. The seeds in slices or chunks of watermelon sold in retail stores or salad bars are a nuisance. One reason that seedless grapes are more popular with consumers than seeded varieties is that the consumer does not have to be concerned with and inconvenienced by the seeds while the fruit is being eaten. With proper care, seedless watermelons have a longer shelf life than seeded melons. This may be due to the fact that flesh Plate 1. break-down occurs in the vicinity of seeds, which are (3N) plants which result from crossing a normal absent, in seedless melons. diploid (2N) plant with a tetraploid (4N). The Hybrid seedless (triploid) watermelons have tetraploid is used as the female or seed parent and the been grown for over 40 years in the United States. diploid is the male or pollen parent ( Figure 1 ). As However, it was not until recently that improved shown by the schematic drawing within figure 1, varieties, aggressive marketing, and increased several steps are necessary in triploid watermelon consumer demand created a rapidly expanding seed production: a diploid (2N) female parent plant is market for seedless watermelons. The seedless treated with colchicine to produce the solid-colored condition is actually sterility resulting from a cross female tetraploid (4N) parent; this is corssed with a between two plants of incompatible chromosome striped male parent (2N) which results in triploid complements. -
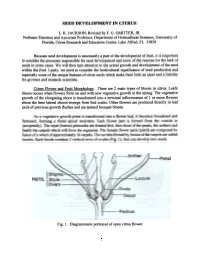
Seed Development in Citrus
SEED DEVELOPMENT IN CITRUS L. K. JACKSON; Revisedby F. G. GMITTER, JR. ProfessorEmeritus and AssociateProfessor, Department of Horticultural Sciences,University of Florida, Citrus Researchand EducationCenter, Lake Alfred, FL 33850 Becauseseed development is necessarilya part of the developmentof fruit. it is important to considerthe processesresponsible for seeddevelopment and someof the reasonsfor the lack of seedsin somecases. We will then turn attentionto the actual growth and developmentof the seed within the fruit. Lastly, we needto considerthe horticultural significanceof seedproduction and especiallysome of the uniquefeatures of citrus seedswhich makethem both an assetand a liability for growersand researchscientists. Citrus Flower and Fruit MoQ1holo~. There are 2 main types of bloom in citrus. Leafy bloom occurswhen flowers form on and with new vegetativegrowth in the spring. The vegetative growth of the elongating shoot is transformedinto a tenninal inflorescenceof 1 or more flowers about the time lateral shootsemerge from bud scales.Other flowers are produceddirectly in leaf axils of previous growth flushesand are termedbouquet bloom. As a vegetative growth point is transfonned into a flower bud, it becomes broadened and flattened, forming a floral apical meristem. Each flower part is fonned from the outside in (acropetally). The sepal (button) primordia are fonned first, then those of the petals, the anthers and finally the carpels which will form the segments. The female flower parts (pistil) are composed by fusion of a whorl of approximately 10 carpels. The cavities fonned by fusion of the carpels are called locules. Each locule contains 2 vertical rows of ovules (Fig. 1), that can develop into seeds. Fig. 1. Diagrammaticportrayal of opencitrus flower.