Microbial Metagenomes and Metatranscriptomes During A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Closest Unicellular Relatives of Animals
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Current Biology, Vol. 12, 1773–1778, October 15, 2002, 2002 Elsevier Science Ltd. All rights reserved. PII S0960-9822(02)01187-9 The Closest Unicellular Relatives of Animals B.F. Lang,1,2 C. O’Kelly,1,3 T. Nerad,4 M.W. Gray,1,5 Results and Discussion and G. Burger1,2,6 1The Canadian Institute for Advanced Research The evolution of the Metazoa from single-celled protists Program in Evolutionary Biology is an issue that has intrigued biologists for more than a 2 De´ partement de Biochimie century. Early morphological and more recent ultra- Universite´ de Montre´ al structural and molecular studies have converged in sup- Succursale Centre-Ville porting the now widely accepted view that animals are Montre´ al, Que´ bec H3C 3J7 related to Fungi, choanoflagellates, and ichthyosporean Canada protists. However, controversy persists as to the spe- 3 Bigelow Laboratory for Ocean Sciences cific evolutionary relationships among these major P.O. Box 475 groups. This uncertainty is reflected in the plethora of 180 McKown Point Road published molecular phylogenies that propose virtually West Boothbay Harbor, Maine 04575 all of the possible alternative tree topologies involving 4 American Type Culture Collection Choanoflagellata, Fungi, Ichthyosporea, and Metazoa. 10801 University Boulevard For example, a monophyletic MetazoaϩChoanoflagel- Manassas, Virginia 20110 lata group has been suggested on the basis of small 5 Department of Biochemistry subunit (SSU) rDNA sequences [1, 6, 7]. Other studies and Molecular Biology using the same sequences have allied Choanoflagellata Dalhousie University with the Fungi [8], placed Choanoflagellata prior to the Halifax, Nova Scotia B3H 4H7 divergence of animals and Fungi [9], or even placed Canada them prior to the divergence of green algae and land plants [10]. -

Sex Is a Ubiquitous, Ancient, and Inherent Attribute of Eukaryotic Life
PAPER Sex is a ubiquitous, ancient, and inherent attribute of COLLOQUIUM eukaryotic life Dave Speijera,1, Julius Lukešb,c, and Marek Eliášd,1 aDepartment of Medical Biochemistry, Academic Medical Center, University of Amsterdam, 1105 AZ, Amsterdam, The Netherlands; bInstitute of Parasitology, Biology Centre, Czech Academy of Sciences, and Faculty of Sciences, University of South Bohemia, 370 05 Ceské Budejovice, Czech Republic; cCanadian Institute for Advanced Research, Toronto, ON, Canada M5G 1Z8; and dDepartment of Biology and Ecology, University of Ostrava, 710 00 Ostrava, Czech Republic Edited by John C. Avise, University of California, Irvine, CA, and approved April 8, 2015 (received for review February 14, 2015) Sexual reproduction and clonality in eukaryotes are mostly Sex in Eukaryotic Microorganisms: More Voyeurs Needed seen as exclusive, the latter being rather exceptional. This view Whereas absence of sex is considered as something scandalous for might be biased by focusing almost exclusively on metazoans. a zoologist, scientists studying protists, which represent the ma- We analyze and discuss reproduction in the context of extant jority of extant eukaryotic diversity (2), are much more ready to eukaryotic diversity, paying special attention to protists. We accept that a particular eukaryotic group has not shown any evi- present results of phylogenetically extended searches for ho- dence of sexual processes. Although sex is very well documented mologs of two proteins functioning in cell and nuclear fusion, in many protist groups, and members of some taxa, such as ciliates respectively (HAP2 and GEX1), providing indirect evidence for (Alveolata), diatoms (Stramenopiles), or green algae (Chlor- these processes in several eukaryotic lineages where sex has oplastida), even serve as models to study various aspects of sex- – not been observed yet. -

Development of Molecular Probes for Dinophysis (Dinophyceae) Plastid: a Tool to Predict Blooming and Explore Plastid Origin
Development of Molecular Probes for Dinophysis (Dinophyceae) Plastid: A Tool to Predict Blooming and Explore Plastid Origin Yoshiaki Takahashi,1 Kiyotaka Takishita,2 Kazuhiko Koike,1 Tadashi Maruyama,2 Takeshi Nakayama,3 Atsushi Kobiyama,1 Takehiko Ogata1 1School of Fisheries Sciences, Kitasato University, Sanriku, Ofunato, Iwate, 022-01011, Japan 2Marine Biotechnology Institute, Heita Kamaishi, Iwate, 026-0001, Japan 3Institute of Biological Sciences, University of Tsukuba, Tennoh-dai, Tsukuba, Ibaraki, 305-8577, Japan Received: 9 July 2004 / Accepted: 19 August 2004 / Online publication: 24 March 2005 Abstract Introduction Dinophysis are species of dinoflagellates that cause Some phytoplankton species are known to produce diarrhetic shellfish poisoning. We have previously toxins that accumulate in plankton feeders. In par- reported that they probably acquire plastids from ticular, toxin accumulation in bivalves causes food cryptophytes in the environment, after which they poisoning in humans, and often leads to severe eco- bloom. Thus monitoring the intracellular plastid nomic damage to the shellfish industry. density in Dinophysis and the source cryptophytes Diarrhetic shellfish poisoning (DSP) is a gastro- occurring in the field should allow prediction of intestinal syndrome caused by phytoplankton tox- Dinophysis blooming. In this study the nucleotide ins, including okadaic acid, and several analogues of sequences of the plastid-encoded small subunit dinophysistoxin (Yasumoto et al., 1985). These tox- ribosomal RNA gene and rbcL (encoding the large ins are derived from several species of dinoflagellates subunit of RuBisCO) from Dinophysis spp. were belonging to the genus Dinophysis (Yasumoto et al, compared with those of cryptophytes, and genetic 1980; Lee et al., 1989). Despite extensive studies in probes specific for the Dinophysis plastid were de- the last 2 decades, little is known about the eco- signed. -
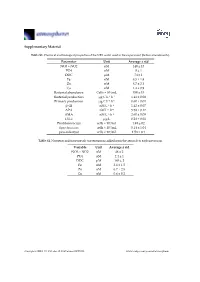
Supplementary Material Parameter Unit Average ± Std NO3 + NO2 Nm
Supplementary Material Table S1. Chemical and biological properties of the NRS water used in the experiment (before amendments). Parameter Unit Average ± std NO3 + NO2 nM 140 ± 13 PO4 nM 8 ± 1 DOC μM 74 ± 1 Fe nM 8.5 ± 1.8 Zn nM 8.7 ± 2.1 Cu nM 1.4 ± 0.9 Bacterial abundance Cells × 104/mL 350 ± 15 Bacterial production μg C L−1 h−1 1.41 ± 0.08 Primary production μg C L−1 h−1 0.60 ± 0.01 β-Gl nM L−1 h−1 1.42 ± 0.07 APA nM L−1 h−1 5.58 ± 0.17 AMA nM L−1·h−1 2.60 ± 0.09 Chl-a μg/L 0.28 ± 0.01 Prochlorococcus cells × 104/mL 1.49 ± 02 Synechococcus cells × 104/mL 5.14 ± 1.04 pico-eukaryot cells × 103/mL 1.58 × 0.1 Table S2. Nutrients and trace metals concentrations added from the aerosols to each mesocosm. Variable Unit Average ± std NO3 + NO2 nM 48 ± 2 PO4 nM 2.4 ± 1 DOC μM 165 ± 2 Fe nM 2.6 ± 1.5 Zn nM 6.7 ± 2.5 Cu nM 0.6 ± 0.2 Atmosphere 2019, 10, 358; doi:10.3390/atmos10070358 www.mdpi.com/journal/atmosphere Atmosphere 2019, 10, 358 2 of 6 Table S3. ANOVA test results between control, ‘UV-treated’ and ‘live-dust’ treatments at 20 h or 44 h, with significantly different values shown in bold. ANOVA df Sum Sq Mean Sq F Value p-value Chl-a 20 H 2, 6 0.03, 0.02 0.02, 0 4.52 0.0634 44 H 2, 6 0.02, 0 0.01, 0 23.13 0.002 Synechococcus Abundance 20 H 2, 7 8.23 × 107, 4.11 × 107 4.11 × 107, 4.51 × 107 0.91 0.4509 44 H 2, 7 5.31 × 108, 6.97 × 107 2.65 × 108, 1.16 × 107 22.84 0.0016 Prochlorococcus Abundance 20 H 2, 8 4.22 × 107, 2.11 × 107 2.11 × 107, 2.71 × 106 7.77 0.0216 44 H 2, 8 9.02 × 107, 1.47 × 107 4.51 × 107, 2.45 × 106 18.38 0.0028 Pico-eukaryote -

Ocean Acidification and High Irradiance Stimulate Growth of The
Biogeosciences Discuss., https://doi.org/10.5194/bg-2019-97 Manuscript under review for journal Biogeosciences Discussion started: 26 March 2019 c Author(s) 2019. CC BY 4.0 License. Ocean acidification and high irradiance stimulate growth of the Antarctic cryptophyte Geminigera cryophila Scarlett Trimborn1,2, Silke Thoms1, Pascal Karitter1, Kai Bischof2 5 1EcoTrace, Biogeosciences section, Alfred Wegener Institute, Bremerhaven, 27568, Germany 2Marine Botany, Department 2 Biology/Chemistry, University of Bremen, Bremen, 28359, Germany Correspondence to: Scarlett Trimborn ([email protected]) Abstract. Ecophysiological studies on Antarctic cryptophytes to assess whether climatic changes such as ocean acidification 10 and enhanced stratification affect their growth in Antarctic coastal waters in the future are lacking so far. This is the first study that investigated the combined effects of increasing availability of pCO2 (400 and 1000 µatm) and irradiance (20, 200 and 500 μmol photons m−2 s−1) on growth, elemental composition and photophysiology of the Antarctic cryptophyte Geminigera cryophila. Under ambient pCO2, this species was characterized by a pronounced sensitivity to increasing irradiance with complete growth inhibition at the highest light intensity. Interestingly, when 15 grown under high pCO2 this negative light effect vanished and it reached highest rates of growth and particulate organic carbon production at the highest irradiance compared to the other tested experimental conditions. Our results for G. cryophila reveal beneficial effects of ocean acidification in conjunction with enhanced irradiance on growth and photosynthesis. Hence, cryptophytes such as G. cryophila may be potential winners of climate change, potentially thriving better in more stratified and acidic coastal waters and contributing in higher abundance to future 20 phytoplankton assemblages of coastal Antarctic waters. -

Predatory Flagellates – the New Recently Discovered Deep Branches of the Eukaryotic Tree and Their Evolutionary and Ecological Significance
Protistology 14 (1), 15–22 (2020) Protistology Predatory flagellates – the new recently discovered deep branches of the eukaryotic tree and their evolutionary and ecological significance Denis V. Tikhonenkov Papanin Institute for Biology of Inland Waters, Russian Academy of Sciences, Borok, 152742, Russia | Submitted March 20, 2020 | Accepted April 6, 2020 | Summary Predatory protists are poorly studied, although they are often representing important deep-branching evolutionary lineages and new eukaryotic supergroups. This short review/opinion paper is inspired by the recent discoveries of various predatory flagellates, which form sister groups of the giant eukaryotic clusters on phylogenetic trees, and illustrate an ancestral state of one or another supergroup of eukaryotes. Here we discuss their evolutionary and ecological relevance and show that the study of such protists may be essential in addressing previously puzzling evolutionary problems, such as the origin of multicellular animals, the plastid spread trajectory, origins of photosynthesis and parasitism, evolution of mitochondrial genomes. Key words: evolution of eukaryotes, heterotrophic flagellates, mitochondrial genome, origin of animals, photosynthesis, predatory protists, tree of life Predatory flagellates and diversity of eu- of the hidden diversity of protists (Moon-van der karyotes Staay et al., 2000; López-García et al., 2001; Edg- comb et al., 2002; Massana et al., 2004; Richards The well-studied multicellular animals, plants and Bass, 2005; Tarbe et al., 2011; de Vargas et al., and fungi immediately come to mind when we hear 2015). In particular, several prevailing and very abun- the term “eukaryotes”. However, these groups of dant ribogroups such as MALV, MAST, MAOP, organisms represent a minority in the real diversity MAFO (marine alveolates, stramenopiles, opistho- of evolutionary lineages of eukaryotes. -

Ciliate Diversity, Community Structure, and Novel Taxa in Lakes of the Mcmurdo Dry Valleys, Antarctica
Reference: Biol. Bull. 227: 175–190. (October 2014) © 2014 Marine Biological Laboratory Ciliate Diversity, Community Structure, and Novel Taxa in Lakes of the McMurdo Dry Valleys, Antarctica YUAN XU1,*†, TRISTA VICK-MAJORS2, RACHAEL MORGAN-KISS3, JOHN C. PRISCU2, AND LINDA AMARAL-ZETTLER4,5,* 1Laboratory of Protozoology, Institute of Evolution & Marine Biodiversity, Ocean University of China, Qingdao 266003, China; 2Montana State University, Department of Land Resources and Environmental Sciences, 334 Leon Johnson Hall, Bozeman, Montana 59717; 3Department of Microbiology, Miami University, Oxford, Ohio 45056; 4The Josephine Bay Paul Center for Comparative Molecular Biology and Evolution, Marine Biological Laboratory, Woods Hole, Massachusetts 02543; and 5Department of Earth, Environmental and Planetary Sciences, Brown University, Providence, Rhode Island 02912 Abstract. We report an in-depth survey of next-genera- trends in dissolved oxygen concentration and salinity may tion DNA sequencing of ciliate diversity and community play a critical role in structuring ciliate communities. A structure in two permanently ice-covered McMurdo Dry PCR-based strategy capitalizing on divergent eukaryotic V9 Valley lakes during the austral summer and autumn (No- hypervariable region ribosomal RNA gene targets unveiled vember 2007 and March 2008). We tested hypotheses on the two new genera in these lakes. A novel taxon belonging to relationship between species richness and environmental an unknown class most closely related to Cryptocaryon conditions -

Protist Phylogeny and the High-Level Classification of Protozoa
Europ. J. Protistol. 39, 338–348 (2003) © Urban & Fischer Verlag http://www.urbanfischer.de/journals/ejp Protist phylogeny and the high-level classification of Protozoa Thomas Cavalier-Smith Department of Zoology, University of Oxford, South Parks Road, Oxford, OX1 3PS, UK; E-mail: [email protected] Received 1 September 2003; 29 September 2003. Accepted: 29 September 2003 Protist large-scale phylogeny is briefly reviewed and a revised higher classification of the kingdom Pro- tozoa into 11 phyla presented. Complementary gene fusions reveal a fundamental bifurcation among eu- karyotes between two major clades: the ancestrally uniciliate (often unicentriolar) unikonts and the an- cestrally biciliate bikonts, which undergo ciliary transformation by converting a younger anterior cilium into a dissimilar older posterior cilium. Unikonts comprise the ancestrally unikont protozoan phylum Amoebozoa and the opisthokonts (kingdom Animalia, phylum Choanozoa, their sisters or ancestors; and kingdom Fungi). They share a derived triple-gene fusion, absent from bikonts. Bikonts contrastingly share a derived gene fusion between dihydrofolate reductase and thymidylate synthase and include plants and all other protists, comprising the protozoan infrakingdoms Rhizaria [phyla Cercozoa and Re- taria (Radiozoa, Foraminifera)] and Excavata (phyla Loukozoa, Metamonada, Euglenozoa, Percolozoa), plus the kingdom Plantae [Viridaeplantae, Rhodophyta (sisters); Glaucophyta], the chromalveolate clade, and the protozoan phylum Apusozoa (Thecomonadea, Diphylleida). Chromalveolates comprise kingdom Chromista (Cryptista, Heterokonta, Haptophyta) and the protozoan infrakingdom Alveolata [phyla Cilio- phora and Miozoa (= Protalveolata, Dinozoa, Apicomplexa)], which diverged from a common ancestor that enslaved a red alga and evolved novel plastid protein-targeting machinery via the host rough ER and the enslaved algal plasma membrane (periplastid membrane). -

Protistology an International Journal Vol
Protistology An International Journal Vol. 10, Number 2, 2016 ___________________________________________________________________________________ CONTENTS INTERNATIONAL SCIENTIFIC FORUM «PROTIST–2016» Yuri Mazei (Vice-Chairman) Welcome Address 2 Organizing Committee 3 Organizers and Sponsors 4 Abstracts 5 Author Index 94 Forum “PROTIST-2016” June 6–10, 2016 Moscow, Russia Website: http://onlinereg.ru/protist-2016 WELCOME ADDRESS Dear colleagues! Republic) entitled “Diplonemids – new kids on the block”. The third lecture will be given by Alexey The Forum “PROTIST–2016” aims at gathering Smirnov (Saint Petersburg State University, Russia): the researchers in all protistological fields, from “Phylogeny, diversity, and evolution of Amoebozoa: molecular biology to ecology, to stimulate cross- new findings and new problems”. Then Sandra disciplinary interactions and establish long-term Baldauf (Uppsala University, Sweden) will make a international scientific cooperation. The conference plenary presentation “The search for the eukaryote will cover a wide range of fundamental and applied root, now you see it now you don’t”, and the fifth topics in Protistology, with the major focus on plenary lecture “Protist-based methods for assessing evolution and phylogeny, taxonomy, systematics and marine water quality” will be made by Alan Warren DNA barcoding, genomics and molecular biology, (Natural History Museum, United Kingdom). cell biology, organismal biology, parasitology, diversity and biogeography, ecology of soil and There will be two symposia sponsored by ISoP: aquatic protists, bioindicators and palaeoecology. “Integrative co-evolution between mitochondria and their hosts” organized by Sergio A. Muñoz- The Forum is organized jointly by the International Gómez, Claudio H. Slamovits, and Andrew J. Society of Protistologists (ISoP), International Roger, and “Protists of Marine Sediments” orga- Society for Evolutionary Protistology (ISEP), nized by Jun Gong and Virginia Edgcomb. -

S41467-021-25308-W.Pdf
ARTICLE https://doi.org/10.1038/s41467-021-25308-w OPEN Phylogenomics of a new fungal phylum reveals multiple waves of reductive evolution across Holomycota ✉ ✉ Luis Javier Galindo 1 , Purificación López-García 1, Guifré Torruella1, Sergey Karpov2,3 & David Moreira 1 Compared to multicellular fungi and unicellular yeasts, unicellular fungi with free-living fla- gellated stages (zoospores) remain poorly known and their phylogenetic position is often 1234567890():,; unresolved. Recently, rRNA gene phylogenetic analyses of two atypical parasitic fungi with amoeboid zoospores and long kinetosomes, the sanchytrids Amoeboradix gromovi and San- chytrium tribonematis, showed that they formed a monophyletic group without close affinity with known fungal clades. Here, we sequence single-cell genomes for both species to assess their phylogenetic position and evolution. Phylogenomic analyses using different protein datasets and a comprehensive taxon sampling result in an almost fully-resolved fungal tree, with Chytridiomycota as sister to all other fungi, and sanchytrids forming a well-supported, fast-evolving clade sister to Blastocladiomycota. Comparative genomic analyses across fungi and their allies (Holomycota) reveal an atypically reduced metabolic repertoire for sanchy- trids. We infer three main independent flagellum losses from the distribution of over 60 flagellum-specific proteins across Holomycota. Based on sanchytrids’ phylogenetic position and unique traits, we propose the designation of a novel phylum, Sanchytriomycota. In addition, our results indicate that most of the hyphal morphogenesis gene repertoire of multicellular fungi had already evolved in early holomycotan lineages. 1 Ecologie Systématique Evolution, CNRS, Université Paris-Saclay, AgroParisTech, Orsay, France. 2 Zoological Institute, Russian Academy of Sciences, St. ✉ Petersburg, Russia. 3 St. -

Phylogenomics Supports the Monophyly of the Cercozoa T ⁎ Nicholas A.T
Molecular Phylogenetics and Evolution 130 (2019) 416–423 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Phylogenomics supports the monophyly of the Cercozoa T ⁎ Nicholas A.T. Irwina, , Denis V. Tikhonenkova,b, Elisabeth Hehenbergera,1, Alexander P. Mylnikovb, Fabien Burkia,2, Patrick J. Keelinga a Department of Botany, University of British Columbia, Vancouver V6T 1Z4, British Columbia, Canada b Institute for Biology of Inland Waters, Russian Academy of Sciences, Borok 152742, Russia ARTICLE INFO ABSTRACT Keywords: The phylum Cercozoa consists of a diverse assemblage of amoeboid and flagellated protists that forms a major Cercozoa component of the supergroup, Rhizaria. However, despite its size and ubiquity, the phylogeny of the Cercozoa Rhizaria remains unclear as morphological variability between cercozoan species and ambiguity in molecular analyses, Phylogeny including phylogenomic approaches, have produced ambiguous results and raised doubts about the monophyly Phylogenomics of the group. Here we sought to resolve these ambiguities using a 161-gene phylogenetic dataset with data from Single-cell transcriptomics newly available genomes and deeply sequenced transcriptomes, including three new transcriptomes from Aurigamonas solis, Abollifer prolabens, and a novel species, Lapot gusevi n. gen. n. sp. Our phylogenomic analysis strongly supported a monophyletic Cercozoa, and approximately-unbiased tests rejected the paraphyletic topologies observed in previous studies. The transcriptome of L. gusevi represents the first transcriptomic data from the large and recently characterized Aquavolonidae-Treumulida-'Novel Clade 12′ group, and phyloge- nomics supported its position as sister to the cercozoan subphylum, Endomyxa. These results provide insights into the phylogeny of the Cercozoa and the Rhizaria as a whole. -

A Distinct Lineage of Giant Viruses Brings a Rhodopsin Photosystem to Unicellular Marine Predators
A distinct lineage of giant viruses brings a rhodopsin photosystem to unicellular marine predators David M. Needhama,1, Susumu Yoshizawab,1, Toshiaki Hosakac,1, Camille Poiriera,d, Chang Jae Choia,d, Elisabeth Hehenbergera,d, Nicholas A. T. Irwine, Susanne Wilkena,2, Cheuk-Man Yunga,d, Charles Bachya,3, Rika Kuriharaf, Yu Nakajimab, Keiichi Kojimaf, Tomomi Kimura-Someyac, Guy Leonardg, Rex R. Malmstromh, Daniel R. Mendei, Daniel K. Olsoni, Yuki Sudof, Sebastian Sudeka, Thomas A. Richardsg, Edward F. DeLongi, Patrick J. Keelinge, Alyson E. Santoroj, Mikako Shirouzuc, Wataru Iwasakib,k,4, and Alexandra Z. Wordena,d,4 aMonterey Bay Aquarium Research Institute, Moss Landing, CA 95039; bAtmosphere & Ocean Research Institute, University of Tokyo, Chiba 277-8564, Japan; cLaboratory for Protein Functional & Structural Biology, RIKEN Center for Biosystems Dynamics Research, Yokohama, Kanagawa 230-0045, Japan; dOcean EcoSystems Biology Unit, GEOMAR Helmholtz Centre for Ocean Research, 24105 Kiel, Germany; eDepartment of Botany, University of British Columbia, Vancouver, BC V6T 1Z4, Canada; fGraduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, Okayama 700-8530, Japan; gLiving Systems Institute, School of Biosciences, College of Life and Environmental Sciences, University of Exeter, Exeter EX4 4SB, United Kingdom; hDepartment of Energy Joint Genome Institute, Walnut Creek, CA 94598; iDaniel K. Inouye Center for Microbial Oceanography, University of Hawaii, Manoa, Honolulu, HI 96822; jDepartment of Ecology, Evolution and Marine Biology, University of California, Santa Barbara, CA 93106; and kDepartment of Biological Sciences, Graduate School of Science, University of Tokyo, Tokyo 113-0032, Japan Edited by W. Ford Doolittle, Dalhousie University, Halifax, Canada, and approved August 8, 2019 (received for review May 27, 2019) Giant viruses are remarkable for their large genomes, often rivaling genomes that range up to 2.4 Mb (Fig.