Green Algae and the Origin of Land Plants1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Early Photosynthetic Eukaryotes Inhabited Low-Salinity Habitats
Early photosynthetic eukaryotes inhabited PNAS PLUS low-salinity habitats Patricia Sánchez-Baracaldoa,1, John A. Ravenb,c, Davide Pisanid,e, and Andrew H. Knollf aSchool of Geographical Sciences, University of Bristol, Bristol BS8 1SS, United Kingdom; bDivision of Plant Science, University of Dundee at the James Hutton Institute, Dundee DD2 5DA, United Kingdom; cPlant Functional Biology and Climate Change Cluster, University of Technology Sydney, Ultimo, NSW 2007, Australia; dSchool of Biological Sciences, University of Bristol, Bristol BS8 1TH, United Kingdom; eSchool of Earth Sciences, University of Bristol, Bristol BS8 1TH, United Kingdom; and fDepartment of Organismic and Evolutionary Biology, Harvard University, Cambridge, MA 02138 Edited by Peter R. Crane, Oak Spring Garden Foundation, Upperville, Virginia, and approved July 7, 2017 (received for review December 7, 2016) The early evolutionary history of the chloroplast lineage remains estimates for the origin of plastids ranging over 800 My (7). At the an open question. It is widely accepted that the endosymbiosis that same time, the ecological setting in which this endosymbiotic event established the chloroplast lineage in eukaryotes can be traced occurred has not been fully explored (8), partly because of phy- back to a single event, in which a cyanobacterium was incorpo- logenetic uncertainties and preservational biases of the fossil re- rated into a protistan host. It is still unclear, however, which cord. Phylogenomics and trait evolution analysis have pointed to a Cyanobacteria are most closely related to the chloroplast, when the freshwater origin for Cyanobacteria (9–11), providing an approach plastid lineage first evolved, and in what habitats this endosym- to address the early diversification of terrestrial biota for which the biotic event occurred. -

Universidad Autónoma De Nuevo León Facultad De Ciencias Biológicas
UNIVERSIDAD AUTÓNOMA DE NUEVO LEÓN FACULTAD DE CIENCIAS BIOLÓGICAS TESIS TAXONOMÍA, DISTRIBUCIÓN E IMPORTANCIA DE LAS ALGAS DE NUEVO LEÓN POR DIANA ELENA AGUIRRE CAVAZOS COMO REQUISITO PARCIAL PARA OBTENER EL GRADO DE DOCTOR EN CIENCIAS CON ACENTUACIÓN EN MANEJO Y ADMINISTRACIÓN DE RECURSOS VEGETALES MAYO, 2018 TAXONOMÍA, DISTRIBUCIÓN E IMPORTANCIA DE LAS ALGAS DE NUEVO LEÓN Comité de Tesis Presidente: Dr. Sergio Manuel Salcedo Martínez. Secretario: Dr. Sergio Moreno Limón. Vocal 1: Hugo Alberto Luna Olvera. Vocal 2: Dr. Marco Antonio Alvarado Vázquez. Vocal 3: Dra. Alejandra Rocha Estrada. TAXONOMÍA, DISTRIBUCIÓN E IMPORTANCIA DE LAS ALGAS DE NUEVO LEÓN Dirección de Tesis Director: Dr. Sergio Manuel Salcedo Martínez. AGRADECIMIENTOS A Dios, por guiar siempre mis pasos y darme fortaleza ante las dificultades. Al Dr. Sergio Manuel Salcedo Martínez, por su disposición para participar como director de este proyecto, por sus consejos y enseñanzas que siempre tendré presente tanto en mi vida profesional como personal; pero sobre todo por su dedicación, paciencia y comprensión que hicieron posible la realización de este trabajo. A la Dra. Alejandra Rocha Estrada, El Dr. Marco Antonio Alvarado Vázquez, el Dr. Sergio Moreno Limón y el Dr. Hugo Alberto Luna Olvera por su apoyo y aportaciones para la realización de este trabajo. Al Dr. Eberto Novelo, por sus valiosas aportaciones para enriquecer el listado taxonómico. A la M.C. Cecilia Galicia Campos, gracias Cecy, por hacer tan amena la estancia en el laboratorio y en el Herbario; por esas pláticas interminables y esas “riso terapias” que siempre levantaban el ánimo. A mis entrañables amigos, “los biólogos”, “los cacos”: Brenda, Libe, Lula, Samy, David, Gera, Pancho, Reynaldo y Ricardo. -

The Lichens' Microbiota, Still a Mystery?
fmicb-12-623839 March 24, 2021 Time: 15:25 # 1 REVIEW published: 30 March 2021 doi: 10.3389/fmicb.2021.623839 The Lichens’ Microbiota, Still a Mystery? Maria Grimm1*, Martin Grube2, Ulf Schiefelbein3, Daniela Zühlke1, Jörg Bernhardt1 and Katharina Riedel1 1 Institute of Microbiology, University Greifswald, Greifswald, Germany, 2 Institute of Plant Sciences, Karl-Franzens-University Graz, Graz, Austria, 3 Botanical Garden, University of Rostock, Rostock, Germany Lichens represent self-supporting symbioses, which occur in a wide range of terrestrial habitats and which contribute significantly to mineral cycling and energy flow at a global scale. Lichens usually grow much slower than higher plants. Nevertheless, lichens can contribute substantially to biomass production. This review focuses on the lichen symbiosis in general and especially on the model species Lobaria pulmonaria L. Hoffm., which is a large foliose lichen that occurs worldwide on tree trunks in undisturbed forests with long ecological continuity. In comparison to many other lichens, L. pulmonaria is less tolerant to desiccation and highly sensitive to air pollution. The name- giving mycobiont (belonging to the Ascomycota), provides a protective layer covering a layer of the green-algal photobiont (Dictyochloropsis reticulata) and interspersed cyanobacterial cell clusters (Nostoc spec.). Recently performed metaproteome analyses Edited by: confirm the partition of functions in lichen partnerships. The ample functional diversity Nathalie Connil, Université de Rouen, France of the mycobiont contrasts the predominant function of the photobiont in production Reviewed by: (and secretion) of energy-rich carbohydrates, and the cyanobiont’s contribution by Dirk Benndorf, nitrogen fixation. In addition, high throughput and state-of-the-art metagenomics and Otto von Guericke University community fingerprinting, metatranscriptomics, and MS-based metaproteomics identify Magdeburg, Germany Guilherme Lanzi Sassaki, the bacterial community present on L. -

Akashiwo Sanguinea
Ocean ORIGINAL ARTICLE and Coastal http://doi.org/10.1590/2675-2824069.20-004hmdja Research ISSN 2675-2824 Phytoplankton community in a tropical estuarine gradient after an exceptional harmful bloom of Akashiwo sanguinea (Dinophyceae) in the Todos os Santos Bay Helen Michelle de Jesus Affe1,2,* , Lorena Pedreira Conceição3,4 , Diogo Souza Bezerra Rocha5 , Luis Antônio de Oliveira Proença6 , José Marcos de Castro Nunes3,4 1 Universidade do Estado do Rio de Janeiro - Faculdade de Oceanografia (Bloco E - 900, Pavilhão João Lyra Filho, 4º andar, sala 4018, R. São Francisco Xavier, 524 - Maracanã - 20550-000 - Rio de Janeiro - RJ - Brazil) 2 Instituto Nacional de Pesquisas Espaciais/INPE - Rede Clima - Sub-rede Oceanos (Av. dos Astronautas, 1758. Jd. da Granja -12227-010 - São José dos Campos - SP - Brazil) 3 Universidade Estadual de Feira de Santana - Departamento de Ciências Biológicas - Programa de Pós-graduação em Botânica (Av. Transnordestina s/n - Novo Horizonte - 44036-900 - Feira de Santana - BA - Brazil) 4 Universidade Federal da Bahia - Instituto de Biologia - Laboratório de Algas Marinhas (Rua Barão de Jeremoabo, 668 - Campus de Ondina 40170-115 - Salvador - BA - Brazil) 5 Instituto Internacional para Sustentabilidade - (Estr. Dona Castorina, 124 - Jardim Botânico - 22460-320 - Rio de Janeiro - RJ - Brazil) 6 Instituto Federal de Santa Catarina (Av. Ver. Abrahão João Francisco, 3899 - Ressacada, Itajaí - 88307-303 - SC - Brazil) * Corresponding author: [email protected] ABSTRAct The objective of this study was to evaluate variations in the composition and abundance of the phytoplankton community after an exceptional harmful bloom of Akashiwo sanguinea that occurred in Todos os Santos Bay (BTS) in early March, 2007. -

Perspectives in Phycology Vol
Perspectives in Phycology Vol. 3 (2016), Issue 3, p. 141–154 Article Published online June 2016 Diversity and ecology of green microalgae in marine systems: an overview based on 18S rRNA gene sequences Margot Tragin1, Adriana Lopes dos Santos1, Richard Christen2,3 and Daniel Vaulot1* 1 Sorbonne Universités, UPMC Univ Paris 06, CNRS, UMR 7144, Station Biologique, Place Georges Teissier, 29680 Roscoff, France 2 CNRS, UMR 7138, Systématique Adaptation Evolution, Parc Valrose, BP71. F06108 Nice cedex 02, France 3 Université de Nice-Sophia Antipolis, UMR 7138, Systématique Adaptation Evolution, Parc Valrose, BP71. F06108 Nice cedex 02, France * Corresponding author: [email protected] With 5 figures in the text and an electronic supplement Abstract: Green algae (Chlorophyta) are an important group of microalgae whose diversity and ecological importance in marine systems has been little studied. In this review, we first present an overview of Chlorophyta taxonomy and detail the most important groups from the marine environment. Then, using public 18S rRNA Chlorophyta sequences from culture and natural samples retrieved from the annotated Protist Ribosomal Reference (PR²) database, we illustrate the distribution of different green algal lineages in the oceans. The largest group of sequences belongs to the class Mamiellophyceae and in particular to the three genera Micromonas, Bathycoccus and Ostreococcus. These sequences originate mostly from coastal regions. Other groups with a large number of sequences include the Trebouxiophyceae, Chlorophyceae, Chlorodendrophyceae and Pyramimonadales. Some groups, such as the undescribed prasinophytes clades VII and IX, are mostly composed of environmental sequences. The 18S rRNA sequence database we assembled and validated should be useful for the analysis of metabarcode datasets acquired using next generation sequencing. -

Algae & Marine Plants of Point Reyes
Algae & Marine Plants of Point Reyes Green Algae or Chlorophyta Genus/Species Common Name Acrosiphonia coalita Green rope, Tangled weed Blidingia minima Blidingia minima var. vexata Dwarf sea hair Bryopsis corticulans Cladophora columbiana Green tuft alga Codium fragile subsp. californicum Sea staghorn Codium setchellii Smooth spongy cushion, Green spongy cushion Trentepohlia aurea Ulva californica Ulva fenestrata Sea lettuce Ulva intestinalis Sea hair, Sea lettuce, Gutweed, Grass kelp Ulva linza Ulva taeniata Urospora sp. Brown Algae or Ochrophyta Genus/Species Common Name Alaria marginata Ribbon kelp, Winged kelp Analipus japonicus Fir branch seaweed, Sea fir Coilodesme californica Dactylosiphon bullosus Desmarestia herbacea Desmarestia latifrons Egregia menziesii Feather boa Fucus distichus Bladderwrack, Rockweed Haplogloia andersonii Anderson's gooey brown Laminaria setchellii Southern stiff-stiped kelp Laminaria sinclairii Leathesia marina Sea cauliflower Melanosiphon intestinalis Twisted sea tubes Nereocystis luetkeana Bull kelp, Bullwhip kelp, Bladder wrack, Edible kelp, Ribbon kelp Pelvetiopsis limitata Petalonia fascia False kelp Petrospongium rugosum Phaeostrophion irregulare Sand-scoured false kelp Pterygophora californica Woody-stemmed kelp, Stalked kelp, Walking kelp Ralfsia sp. Silvetia compressa Rockweed Stephanocystis osmundacea Page 1 of 4 Red Algae or Rhodophyta Genus/Species Common Name Ahnfeltia fastigiata Bushy Ahnfelt's seaweed Ahnfeltiopsis linearis Anisocladella pacifica Bangia sp. Bossiella dichotoma Bossiella -

Phylogenetic Placement of Botryococcus Braunii (Trebouxiophyceae) and Botryococcus Sudeticus Isolate Utex 2629 (Chlorophyceae)1
J. Phycol. 40, 412–423 (2004) r 2004 Phycological Society of America DOI: 10.1046/j.1529-8817.2004.03173.x PHYLOGENETIC PLACEMENT OF BOTRYOCOCCUS BRAUNII (TREBOUXIOPHYCEAE) AND BOTRYOCOCCUS SUDETICUS ISOLATE UTEX 2629 (CHLOROPHYCEAE)1 Hoda H. Senousy, Gordon W. Beakes, and Ethan Hack2 School of Biology, University of Newcastle upon Tyne, Newcastle upon Tyne NE1 7RU, UK The phylogenetic placement of four isolates of a potential source of renewable energy in the form of Botryococcus braunii Ku¨tzing and of Botryococcus hydrocarbon fuels (Metzger et al. 1991, Metzger and sudeticus Lemmermann isolate UTEX 2629 was Largeau 1999, Banerjee et al. 2002). The best known investigated using sequences of the nuclear small species is Botryococcus braunii Ku¨tzing. This organism subunit (18S) rRNA gene. The B. braunii isolates has a worldwide distribution in fresh and brackish represent the A (two isolates), B, and L chemical water and is occasionally found in salt water. Although races. One isolate of B. braunii (CCAP 807/1; A race) it grows relatively slowly, it sometimes forms massive has a group I intron at Escherichia coli position 1046 blooms (Metzger et al. 1991, Tyson 1995). Botryococcus and isolate UTEX 2629 has group I introns at E. coli braunii strains differ in the hydrocarbons that they positions 516 and 1512. The rRNA sequences were accumulate, and they have been classified into three aligned with 53 previously reported rRNA se- chemical races, called A, B, and L. Strains in the A race quences from members of the Chlorophyta, includ- accumulate alkadienes; strains in the B race accumulate ing one reported for B. -

Evolution and Comparative Morphology of the Euglenophyte
EVOLUTION AND COMPARATIVE MORPHOLOGY OF THE EUGLENOPHYTE PLASTID by PATRICK JERRY PAUL BROWN (Under the Direction of Mark A. Farmer) ABSTRACT My doctoral research centered on understanding the evolution of the euglenophyte protists, with special attention paid to their plastids. The euglenophytes are a widely distributed group of euglenid protists that have acquired a chloroplast via secondary symbiogenesis. The goals of my research were to 1) test the efficacy of plastid morphological and ultrastructural characters in phylogenetic analysis; 2) understand the process of plastid development and partitioning in the euglenophytes; 3) to use a plastid- encoded protein gene to determine a euglenophyte phylogeny; and 4) to perform a multi- gene analysis to uncover clues about the origins of the euglenophyte plastid. My work began with an alpha-taxonomic study that redefined the rare euglenophyte Euglena rustica. This work not only validly circumscribed the species, but also noted novel features of its habitat, cyclic migration habits, and cellular biology. This was followed by a study of plastid morphology and development in a number of diverse euglenophytes. The results of this study showed that the plastids of euglenophytes undergo drastic changes in morphology and ultrastructure over the course of a single cell division cycle. I concluded that there are four main classes of plastid development and partitioning in the euglenophytes, and that the class a given species will use is dependant on its interphase plastid morphology and the rigidity of the cell. The discovery of the class IV partitioning strategy in which cells with only one or very few plastids fragment their plastids prior to cell division was very significant. -

Genes in Some Samples, Suggesting That the Gene Repertoire Is Modulated by Environmental Conditions
Analyses de variations génomiques liées à la biogéographie des picoalgues Mamiellales. Thèse de doctorat de l'université Paris-Saclay École doctorale n°577, Structure et dynamique des systèmes vivants (SDSV) Spécialité de doctorat : Sciences de la Vie et de la Santé Unité de recherche : Université Paris-Saclay, Univ Evry, CNRS, CEA, Génomique métabolique, 91057, Evry-Courcouronnes, France. Référent : Université d'Évry Val d’Essonne Thèse présentée et soutenue à Évry, le 28/08/2020, par Jade LECONTE Composition du Jury Christophe AMBROISE Professeur des universités, Université Examinateur & Président d’Evry Val d’Essonne (UMR 8071) François-Yves BOUGET Directeur de recherche CNRS, Sorbonne Rapporteur & Examinateur Université (UMR 7232) Frédéric PARTENSKY Directeur de recherche CNRS, Sorbonne Rapporteur & Examinateur Université (UMR 7144) Ian PROBERT Ingénieur de recherche, Sorbonne Examinateur Université Olivier JAILLON Directeur de thèse Directeur de recherche, CEA (UMR 8030) Remerciements Je souhaite remercier en premier lieu mon directeur de thèse, Olivier Jaillon, pour son encadrement, ses conseils et sa compréhension depuis mon arrivée au Genoscope. J’ai appris beaucoup à ses côtés, progressé dans de nombreux domaines, et apprécié partager l’unique bureau sans moquette du troisième étage avec lui. Merci également à Patrick Wincker pour m’avoir donné l’opportunité de travailler au sein du LAGE toutes ces années. J’ai grâce à vous deux eu l’occasion de plonger un peu plus loin dans le monde de l’écologie marine à travers le projet Tara Oceans, et j’en suis plus que reconnaissante. Je tiens également à remercier les membres de mon jury de thèse, à la fois mes rapporteurs François-Yves Bouget et Frédéric Partensky qui ont bien voulu évaluer ce manuscrit, ainsi que Christophe Ambroise et Ian Probert qui ont également volontiers accepté de participer à ma soutenance. -
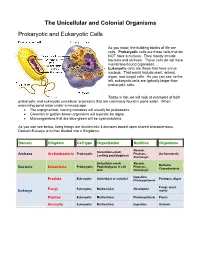
The Unicellular and Colonial Organisms Prokaryotic And
The Unicellular and Colonial Organisms Prokaryotic and Eukaryotic Cells As you know, the building blocks of life are cells. Prokaryotic cells are those cells that do NOT have a nucleus. They mostly include bacteria and archaea. These cells do not have membrane-bound organelles. Eukaryotic cells are those that have a true nucleus. That would include plant, animal, algae, and fungal cells. As you can see, to the left, eukaryotic cells are typically larger than prokaryotic cells. Today in lab, we will look at examples of both prokaryotic and eukaryotic unicellular organisms that are commonly found in pond water. When examining pond water under a microscope… The unpigmented, moving microbes will usually be protozoans. Greenish or golden-brown organisms will typically be algae. Microorganisms that are blue-green will be cyanobacteria. As you can see below, living things are divided into 3 domains based upon shared characteristics. Domain Eukarya is further divided into 4 Kingdoms. Domain Kingdom Cell type Organization Nutrition Organisms Absorb, Unicellular-small; Prokaryotic Photsyn., Archaeacteria Archaea Archaebacteria Lacking peptidoglycan Chemosyn. Unicellular-small; Absorb, Bacteria, Prokaryotic Peptidoglycan in cell Photsyn., Bacteria Eubacteria Cyanobacteria wall Chemosyn. Ingestion, Eukaryotic Unicellular or colonial Protozoa, Algae Protista Photosynthesis Fungi, yeast, Fungi Eukaryotic Multicellular Absorption Eukarya molds Plantae Eukaryotic Multicellular Photosynthesis Plants Animalia Eukaryotic Multicellular Ingestion Animals Prokaryotic Organisms – the archaea, non-photosynthetic bacteria, and cyanobacteria Archaea - Microorganisms that resemble bacteria, but are different from them in certain aspects. Archaea cell walls do not include the macromolecule peptidoglycan, which is always found in the cell walls of bacteria. Archaea usually live in extreme, often very hot or salty environments, such as hot mineral springs or deep-sea hydrothermal vents. -

Lateral Gene Transfer of Anion-Conducting Channelrhodopsins Between Green Algae and Giant Viruses
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.15.042127; this version posted April 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 5 Lateral gene transfer of anion-conducting channelrhodopsins between green algae and giant viruses Andrey Rozenberg 1,5, Johannes Oppermann 2,5, Jonas Wietek 2,3, Rodrigo Gaston Fernandez Lahore 2, Ruth-Anne Sandaa 4, Gunnar Bratbak 4, Peter Hegemann 2,6, and Oded 10 Béjà 1,6 1Faculty of Biology, Technion - Israel Institute of Technology, Haifa 32000, Israel. 2Institute for Biology, Experimental Biophysics, Humboldt-Universität zu Berlin, Invalidenstraße 42, Berlin 10115, Germany. 3Present address: Department of Neurobiology, Weizmann 15 Institute of Science, Rehovot 7610001, Israel. 4Department of Biological Sciences, University of Bergen, N-5020 Bergen, Norway. 5These authors contributed equally: Andrey Rozenberg, Johannes Oppermann. 6These authors jointly supervised this work: Peter Hegemann, Oded Béjà. e-mail: [email protected] ; [email protected] 20 ABSTRACT Channelrhodopsins (ChRs) are algal light-gated ion channels widely used as optogenetic tools for manipulating neuronal activity 1,2. Four ChR families are currently known. Green algal 3–5 and cryptophyte 6 cation-conducting ChRs (CCRs), cryptophyte anion-conducting ChRs (ACRs) 7, and the MerMAID ChRs 8. Here we 25 report the discovery of a new family of phylogenetically distinct ChRs encoded by marine giant viruses and acquired from their unicellular green algal prasinophyte hosts. -

Improved Protocol for the Preparation of Tetraselmis Suecica Axenic
36 The Open Biotechnology Journal, 2010, 4, 36-46 Open Access Improved Protocol for the Preparation of Tetraselmis suecica Axenic Culture and Adaptation to Heterotrophic Cultivation Mojtaba Azma1, Rosfarizan Mohamad1, Raha Abdul Rahim2 and Arbakariya B. Ariff*,1 1Department of Bioprocess Technology, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, 43400 UPM Serdang, Selangor, Malaysia 2Department of Cell and Molecular Biology, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, 43400 UPM Serdang, Selangor, Malaysia Abstract: The effectiveness of various physical and chemical methods for the removal of contaminants from the microal- gae, Tetraselmis suecica, culture was investigated. The information obtained was used as the basis for the development of improved protocol for the preparation of axenic culture to be adapted to heterotrophic cultivation. Repeated centrifugation and rinsing effectively removed the free bacterial contaminants from the microalgae culture while sonication helped to loosen up the tightly attached bacterial contaminants on the microalgae cells. Removal of bacterial spores was accom- plished using a mixture of two antibiotics, 5 mg/mL vancomycine and 10 mg/mL neomycine. Walne medium formulation with natural seawater was preferred for the enhancement of growth of T. suecica. Adaptation of growth from photoautot- rophic to heterotrophic conditions was achieved by the repeated cultivation of photoautotrophic culture with sequential reduction in illumination time, and finally the culture was inoculated into the medium containing 10 g/L glucose, incu- bated in total darkness to obtain heterotrophic cells. Changes in the morphology and composition of T. suecica cells dur- ing the adaptation from photoautotrophic to heterotrophic condition, as examined under Transmission Electron Micro- scope, were also reported.