DNA Damage Checkpoint Dynamics Drive Cell Cycle Phase Transitions
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

DNA Damage Checkpoint Dynamics Drive Cell Cycle Phase Transitions
bioRxiv preprint doi: https://doi.org/10.1101/137307; this version posted August 4, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. DNA damage checkpoint dynamics drive cell cycle phase transitions Hui Xiao Chao1,2, Cere E. Poovey1, Ashley A. Privette1, Gavin D. Grant3,4, Hui Yan Chao1, Jeanette G. Cook3,4, and Jeremy E. Purvis1,2,4,† 1Department of Genetics 2Curriculum for Bioinformatics and Computational Biology 3Department of Biochemistry and Biophysics 4Lineberger Comprehensive Cancer Center University of North Carolina, Chapel Hill 120 Mason Farm Road Chapel Hill, NC 27599-7264 †Corresponding Author: Jeremy Purvis Genetic Medicine Building 5061, CB#7264 120 Mason Farm Road Chapel Hill, NC 27599-7264 [email protected] ABSTRACT DNA damage checkpoints are cellular mechanisms that protect the integrity of the genome during cell cycle progression. In response to genotoxic stress, these checkpoints halt cell cycle progression until the damage is repaired, allowing cells enough time to recover from damage before resuming normal proliferation. Here, we investigate the temporal dynamics of DNA damage checkpoints in individual proliferating cells by observing cell cycle phase transitions following acute DNA damage. We find that in gap phases (G1 and G2), DNA damage triggers an abrupt halt to cell cycle progression in which the duration of arrest correlates with the severity of damage. However, cells that have already progressed beyond a proposed “commitment point” within a given cell cycle phase readily transition to the next phase, revealing a relaxation of checkpoint stringency during later stages of certain cell cycle phases. -

Metabolic Checkpoints in Cancer Cell Cycle
City University of New York (CUNY) CUNY Academic Works All Dissertations, Theses, and Capstone Projects Dissertations, Theses, and Capstone Projects 2-2014 Metabolic Checkpoints in Cancer Cell Cycle Mahesh Saqcena Graduate Center, City University of New York How does access to this work benefit ou?y Let us know! More information about this work at: https://academicworks.cuny.edu/gc_etds/106 Discover additional works at: https://academicworks.cuny.edu This work is made publicly available by the City University of New York (CUNY). Contact: [email protected] METABOLIC CHECKPOINTS IN CANCER CELL CYCLE By Mahesh Saqcena A dissertation submitted to the Graduate Faculty in Biochemistry in partial fulfillment of the requirements for the degree of Doctor of Philosophy, The City University of New York 2014 i 2014 Mahesh Saqcena All Rights Reserved ii This manuscript has been read and accepted for the Graduate Faculty in Biochemistry in satisfaction of the dissertation requirement for the degree of Doctor of Philosophy. Date Dr. David A. Foster (Chair of Examining Committee) Date Dr. Edward J. Kennelly (Executive Officer) Dr. Mitchell Goldfarb (Hunter College) Dr. Paul Feinstein (Hunter College) Dr. Richard Kolesnick (Sloan Kettering Institute) Dr. Frederick R. Cross (Rockefeller University) (Supervisory Committee) THE CITY UNIVERSITY OF NEW YORK iii Abstract METABOLIC CHECKPOINTS IN CANCER CELL CYCLE by Mahesh Saqcena Advisor: Dr. David A. Foster Growth factors (GFs) as well as nutrient sufficiency regulate cell division in metazoans. The vast majority of mutations that contribute to cancer are in genes that regulate progression through the G1 phase of the cell cycle. A key regulatory site in G1 is the growth factor-dependent Restriction Point (R), where cells get permissive signals to divide. -

Mitosis Vs. Meiosis
Mitosis vs. Meiosis In order for organisms to continue growing and/or replace cells that are dead or beyond repair, cells must replicate, or make identical copies of themselves. In order to do this and maintain the proper number of chromosomes, the cells of eukaryotes must undergo mitosis to divide up their DNA. The dividing of the DNA ensures that both the “old” cell (parent cell) and the “new” cells (daughter cells) have the same genetic makeup and both will be diploid, or containing the same number of chromosomes as the parent cell. For reproduction of an organism to occur, the original parent cell will undergo Meiosis to create 4 new daughter cells with a slightly different genetic makeup in order to ensure genetic diversity when fertilization occurs. The four daughter cells will be haploid, or containing half the number of chromosomes as the parent cell. The difference between the two processes is that mitosis occurs in non-reproductive cells, or somatic cells, and meiosis occurs in the cells that participate in sexual reproduction, or germ cells. The Somatic Cell Cycle (Mitosis) The somatic cell cycle consists of 3 phases: interphase, m phase, and cytokinesis. 1. Interphase: Interphase is considered the non-dividing phase of the cell cycle. It is not a part of the actual process of mitosis, but it readies the cell for mitosis. It is made up of 3 sub-phases: • G1 Phase: In G1, the cell is growing. In most organisms, the majority of the cell’s life span is spent in G1. • S Phase: In each human somatic cell, there are 23 pairs of chromosomes; one chromosome comes from the mother and one comes from the father. -

Interferon-A-Induced G1 Phase Arrest Through Up-Regulated Expression of CDK Inhibitors, P19ink4d and P21cip1 in Mouse Macrophages
Oncogene (1998) 16, 2075 ± 2086 1998 Stockton Press All rights reserved 0950 ± 9232/98 $12.00 http://www.stockton-press.co.uk/onc Interferon-a-induced G1 phase arrest through up-regulated expression of CDK inhibitors, p19Ink4D and p21Cip1 in mouse macrophages Masaaki Matsuoka, Kenzaburo Tani and Shigetaka Asano Department of Hematology and Oncology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108, Japan The mechanism of cell cycle arrest induced by interferon-a dependent kinases or cdks (reviewed by Sherr, 1993). (IFN-a) was analysed using a mouse macrophage cell line, D-type cyclins (Lew et al., 1991; Matsushime et al., 1991) BAC1.2F5A. IFN-a added in media before mid-G1 and cyclin E (Ko et al., 1991; Lew et al., 1991) govern the prohibited cells from entering S phase. The blockage of G1/S transition in association with their proper G1/S transition was associated with diminuition of both physiological partners, cdk4 (Matsushime et al., 1992) cyclin D1/cdk4- and cyclin E/cdk2-associated kinase or cdk6 (Meyerson and Harlow, 1994), and cdk2 (Dulic et activities. G1 cyclin-associated kinase activities were al., 1992; Ko et al., 1992), respectively. When cells enter down-regulated quickly after the addition of IFN-a. Cells G1 phase with mitogenic stimuli, the synthesis and the treated with IFN-a contained excess amounts of cdk active complex formation of D-type cyclins and cdk4 are inhibitors which down-regulated G1 cyclin/cdk-associated induced in early to middle G1 phase, and their complex kinase activities in the proliferating cells and this action formation and activities reach maximal levels at late G1 was counteracted by exogenously-supplied recombinant (Matsushime et al., 1992, 1994). -

The Cell Cycle & Mitosis
The Cell Cycle & Mitosis Cell Growth The Cell Cycle is G1 phase ___________________________________ _______________________________ During the Cell Cycle, a cell ___________________________________ ___________________________________ Anaphase Cell Division ___________________________________ Mitosis M phase M ___________________________________ S phase replication DNA Interphase Interphase is ___________________________ ___________________________________ G2 phase Interphase is divided into three phases: ___, ___, & ___ G1 Phase S Phase G2 Phase The G1 phase is a period of The S phase replicates During the G2 phase, many of activity in which cells _______ ________________and the organelles and molecules ____________________ synthesizes _______ molecules. required for ____________ __________ Cells will When DNA replication is ___________________ _______________ and completed, _____________ When G2 is completed, the cell is synthesize new ___________ ____________________ ready to enter the ____________________ ____________________ ____________________ ____________________ ____________________ Mitosis are divided into four phases: _____________, ______________, _____________, & _____________ Below are cells in two different phases of the cell cycle, fill in the blanks using the word bank: Chromatin Nuclear Envelope Chromosome Sister Chromatids Nucleolus Spinder Fiber Centrosome Centrioles 5.._________ 1.__________ v 6..__________ 2.__________ 7.__________ 3.__________ 8..__________ 4.__________ v The Cell Cycle & Mitosis Microscope Lab: -

Loss of P21 Disrupts P14arf-Induced G1 Cell Cycle Arrest but Augments P14arf-Induced Apoptosis in Human Carcinoma Cells
Oncogene (2005) 24, 4114–4128 & 2005 Nature Publishing Group All rights reserved 0950-9232/05 $30.00 www.nature.com/onc Loss of p21 disrupts p14ARF-induced G1 cell cycle arrest but augments p14ARF-induced apoptosis in human carcinoma cells Philipp G Hemmati1,3, Guillaume Normand1,3, Berlinda Verdoodt1, Clarissa von Haefen1, Anne Hasenja¨ ger1, DilekGu¨ ner1, Jana Wendt1, Bernd Do¨ rken1,2 and Peter T Daniel*,1,2 1Department of Hematology, Oncology and Tumor Immunology, University Medical Center Charite´, Campus Berlin-Buch, Berlin-Buch, Germany; 2Max-Delbru¨ck-Center for Molecular Medicine, Berlin-Buch, Germany The human INK4a locus encodes two structurally p16INK4a and p14ARF (termed p19ARF in the mouse), latter unrelated tumor suppressor proteins, p16INK4a and p14ARF of which is transcribed in an Alternative Reading Frame (p19ARF in the mouse), which are frequently inactivated in from a separate exon 1b (Duro et al., 1995; Mao et al., human cancer. Both the proapoptotic and cell cycle- 1995; Quelle et al., 1995; Stone et al., 1995). P14ARF is regulatory functions of p14ARF were initially proposed to usually expressed at low levels, but rapid upregulation be strictly dependent on a functional p53/mdm-2 tumor of p14ARF is triggered by various stimuli, that is, suppressor pathway. However, a number of recent reports the expression of cellular or viral oncogenes including have implicated p53-independent mechanisms in the E2F-1, E1A, c-myc, ras, and v-abl (de Stanchina et al., regulation of cell cycle arrest and apoptosis induction by 1998; Palmero et al., 1998; Radfar et al., 1998; Zindy p14ARF. Here, we show that the G1 cell cycle arrest et al., 1998). -

Regulation of P27kip1 and P57kip2 Functions by Natural Polyphenols
biomolecules Review Regulation of p27Kip1 and p57Kip2 Functions by Natural Polyphenols Gian Luigi Russo 1,* , Emanuela Stampone 2 , Carmen Cervellera 1 and Adriana Borriello 2,* 1 National Research Council, Institute of Food Sciences, 83100 Avellino, Italy; [email protected] 2 Department of Precision Medicine, University of Campania “Luigi Vanvitelli”, 81031 Napoli, Italy; [email protected] * Correspondence: [email protected] (G.L.R.); [email protected] (A.B.); Tel.: +39-0825-299-331 (G.L.R.) Received: 31 July 2020; Accepted: 9 September 2020; Published: 13 September 2020 Abstract: In numerous instances, the fate of a single cell not only represents its peculiar outcome but also contributes to the overall status of an organism. In turn, the cell division cycle and its control strongly influence cell destiny, playing a critical role in targeting it towards a specific phenotype. Several factors participate in the control of growth, and among them, p27Kip1 and p57Kip2, two proteins modulating various transitions of the cell cycle, appear to play key functions. In this review, the major features of p27 and p57 will be described, focusing, in particular, on their recently identified roles not directly correlated with cell cycle modulation. Then, their possible roles as molecular effectors of polyphenols’ activities will be discussed. Polyphenols represent a large family of natural bioactive molecules that have been demonstrated to exhibit promising protective activities against several human diseases. Their use has also been proposed in association with classical therapies for improving their clinical effects and for diminishing their negative side activities. The importance of p27Kip1 and p57Kip2 in polyphenols’ cellular effects will be discussed with the aim of identifying novel therapeutic strategies for the treatment of important human diseases, such as cancers, characterized by an altered control of growth. -

The Cell Cycle Coloring Worksheet
Name: Date: Period: The Cell Cycle Coloring Worksheet Label the diagram below with the following labels: Anaphase Interphase Mitosis Cell division (M Phase) Interphase Prophase Cytokinesis Interphase S-DNA replication G1 – cell grows Metaphase Telophase G2 – prepares for mitosis Then on the diagram, lightly color the G1 phase BLUE, the S phase YELLOW, the G2 phase RED, and the stages of mitosis ORANGE. Color the arrows indicating all of the interphases in GREEN. Color the part of the arrow indicating mitosis PURPLE and the part of the arrow indicating cytokinesis YELLOW. M-PHASE YELLOW: GREEN: CYTOKINESIS INTERPHASE PURPLE: TELOPHASE MITOSIS ANAPHASE ORANGE METAPHASE BLUE: G1: GROWS PROPHASE PURPLE MITOSIS RED:G2: PREPARES GREEN: FOR MITOSIS INTERPHASE YELLOW: S PHASE: DNA REPLICATION GREEN: INTERPHASE Use the diagram and your notes to answer the following questions. 1. What is a series of events that cells go through as they grow and divide? CELL CYCLE 2. What is the longest stage of the cell cycle called? INTERPHASE 3. During what stage does the G1, S, and G2 phases happen? INTERPHASE 4. During what phase of the cell cycle does mitosis and cytokinesis occur? M-PHASE 5. During what phase of the cell cycle does cell division occur? MITOSIS 6. During what phase of the cell cycle is DNA replicated? S-PHASE 7. During what phase of the cell cycle does the cell grow? G1,G2 8. During what phase of the cell cycle does the cell prepare for mitosis? G2 9. How many stages are there in mitosis? 4 10. Put the following stages of mitosis in order: anaphase, prophase, metaphase, and telophase. -

Working on Genomic Stability: from the S-Phase to Mitosis
G C A T T A C G G C A T genes Review Working on Genomic Stability: From the S-Phase to Mitosis Sara Ovejero 1,2,3,* , Avelino Bueno 1,4 and María P. Sacristán 1,4,* 1 Instituto de Biología Molecular y Celular del Cáncer (IBMCC), Universidad de Salamanca-CSIC, Campus Miguel de Unamuno, 37007 Salamanca, Spain; [email protected] 2 Institute of Human Genetics, CNRS, University of Montpellier, 34000 Montpellier, France 3 Department of Biological Hematology, CHU Montpellier, 34295 Montpellier, France 4 Departamento de Microbiología y Genética, Universidad de Salamanca, Campus Miguel de Unamuno, 37007 Salamanca, Spain * Correspondence: [email protected] (S.O.); [email protected] (M.P.S.); Tel.: +34-923-294808 (M.P.S.) Received: 31 January 2020; Accepted: 18 February 2020; Published: 20 February 2020 Abstract: Fidelity in chromosome duplication and segregation is indispensable for maintaining genomic stability and the perpetuation of life. Challenges to genome integrity jeopardize cell survival and are at the root of different types of pathologies, such as cancer. The following three main sources of genomic instability exist: DNA damage, replicative stress, and chromosome segregation defects. In response to these challenges, eukaryotic cells have evolved control mechanisms, also known as checkpoint systems, which sense under-replicated or damaged DNA and activate specialized DNA repair machineries. Cells make use of these checkpoints throughout interphase to shield genome integrity before mitosis. Later on, when the cells enter into mitosis, the spindle assembly checkpoint (SAC) is activated and remains active until the chromosomes are properly attached to the spindle apparatus to ensure an equal segregation among daughter cells. -

S-Phase Entry, Activation of E2F-Responsive Genes, and Apoptosis
Proc. Natl. Acad. Sci. USA Vol. 92, pp. 5436-5440, June 1995 Medical Sciences Deficiency of retinoblastoma protein leads to inappropriate S-phase entry, activation of E2F-responsive genes, and apoptosis (tumor supressor gene/cell cycle control/programmed cell death) ALEXANDRU ALMASAN*tt, YUXIN YIN*t§, RUTH E. KELLY*, EVA Y.-H. P. LEES, ALLAN BRADLEYII, WEIWEI LI**, JOSEPH R. BERTINO**, AND GEOFFREY M. WAHL*tt *Gene Expression Laboratory, The Salk Institute, 10010 North Torrey Pines Road, La Jolla, CA 92037; tCenter for Molecular Medicine and Institute of Biotechnology, The University of Texas Health Science Center at San Antonio, San Antonio, TX 78284; I'Department of Molecular and Human Genetics and Howard Hughes Medical Institute, Baylor College of Medicine, Houston, TX 77030; and **Program for Molecular Pharmacology and Therapeutics, Memorial Sloan-Kettering Cancer Center, New York, NY 10021 Communicated by Leslie E. Orgel, The Salk Institute for Biological Studies, San Diego, CA, January 24, 1995 (received for review December 12, 1994) ABSTRACT The retinoblastoma susceptibility gene (Rb) thymidylate synthase (TS) and other genes necessary for entry participates in controlling the G1/S-phase transition, pre- into and progression through S phase (10, 14). sumably by binding and inactivating E2F transcription acti- The data obtained from studies with viral oncoproteins vator family members. Mouse embryonic fibroblasts (MEFs) suggest that Rb protein acts by sequestering essential factors with no, one, or two inactivated Rb genes were used to determine required for G1/S progression. However, each of these onco- the specific contributions of Rb protein to cell cycle progres- proteins binds other cellular factors such as the Rb-related sion and gene expression. -
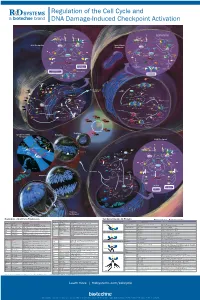
Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation
RnDSy-lu-2945 Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation IR UV IR Stalled Replication Forks/ BRCA1 Rad50 Long Stretches of ss-DNA Rad50 Mre11 BRCA1 Nbs1 Rad9-Rad1-Hus1 Mre11 RPA MDC1 γ-H2AX DNA Pol α/Primase RFC2-5 MDC1 Nbs1 53BP1 MCM2-7 53BP1 γ-H2AX Rad17 Claspin MCM10 Rad9-Rad1-Hus1 TopBP1 CDC45 G1/S Checkpoint Intra-S-Phase RFC2-5 ATM ATR TopBP1 Rad17 ATRIP ATM Checkpoint Claspin Chk2 Chk1 Chk2 Chk1 ATR Rad50 ATRIP Mre11 FANCD2 Ubiquitin MDM2 MDM2 Nbs1 CDC25A Rad50 Mre11 BRCA1 Ub-mediated Phosphatase p53 CDC25A Ubiquitin p53 FANCD2 Phosphatase Degradation Nbs1 p53 p53 CDK2 p21 p21 BRCA1 Ub-mediated SMC1 Degradation Cyclin E/A SMC1 CDK2 Slow S Phase CDC45 Progression p21 DNA Pol α/Primase Slow S Phase p21 Cyclin E Progression Maintenance of Inhibition of New CDC6 CDT1 CDC45 G1/S Arrest Origin Firing ORC MCM2-7 MCM2-7 Recovery of Stalled Replication Forks Inhibition of MCM10 MCM10 Replication Origin Firing DNA Pol α/Primase ORI CDC6 CDT1 MCM2-7 ORC S Phase Delay MCM2-7 MCM10 MCM10 ORI Geminin EGF EGF R GAB-1 CDC6 CDT1 ORC MCM2-7 PI 3-Kinase p70 S6K MCM2-7 S6 Protein Translation Pre-RC (G1) GAB-2 MCM10 GSK-3 TSC1/2 MCM10 ORI PIP2 TOR Promotes Replication CAK EGF Origin Firing Origin PIP3 Activation CDK2 EGF R Akt CDC25A PDK-1 Phosphatase Cyclin E/A SHIP CIP/KIP (p21, p27, p57) (Active) PLCγ PP2A (Active) PTEN CDC45 PIP2 CAK Unwinding RPA CDC7 CDK2 IP3 DAG (Active) Positive DBF4 α Feedback CDC25A DNA Pol /Primase Cyclin E Loop Phosphatase PKC ORC RAS CDK4/6 CDK2 (Active) Cyclin E MCM10 CDC45 RPA IP Receptor -

Cell Cycle Checkpoint in Cancer: a Therapeutically Targetable Double-Edged Sword Roberta Visconti1*, Rosa Della Monica2,3 and Domenico Grieco2,3*
Visconti et al. Journal of Experimental & Clinical Cancer Research (2016) 35:153 DOI 10.1186/s13046-016-0433-9 REVIEW Open Access Cell cycle checkpoint in cancer: a therapeutically targetable double-edged sword Roberta Visconti1*, Rosa Della Monica2,3 and Domenico Grieco2,3* Abstract Major currently used anticancer therapeutics either directly damage DNA or target and upset basic cell division mechanisms like DNA replication and chromosome segregation. These insults elicit activation of cell cycle checkpoints, safeguard mechanisms that cells implement to correctly complete cell cycle phases, repair damage or eventually commit suicide in case damage is unrepairable. Although cancer cells appear to be advantageously defective in some aspects of checkpoint physiology, recent acquisitions on the biochemical mechanisms of the various checkpoints are offering new therapeutic approaches against cancer. Indeed, chemical manipulation of these mechanisms is providing new therapeutic strategies and tools to increase the killing efficacy of major cancer therapeutics as well as to directly promote cancer cell death. In this review we summarize developing concepts on how targeting cell cycle checkpoints may provide substantial improvement to cancer therapy. Keywords: Cell cycle checkpoint, Cancer drug, DNA damage, Mitosis, Chk1, Wee1, Spindle assembly checkpoint, Taxane, Vinca alkaloids, Fcp1 Background opportunities for cancer treatment. Here we will review The mechanisms of cell division and the genome itself how recent efforts to identify new strategies and drugs are routinely endangered by endogenous and exogenous targeting cell cycle checkpoints will likely translate soon insults. For instance reactive oxygen species, produced into benefit to clinical practice in oncology. As outlined during metabolic reactions, inflammation or exciting in Table 1, we will focus our attention on drugs targeting and ionizing radiations, can damage chromosomes and key players of the S and G2/M checkpoints activated in upset chromosome replication and segregation [1, 2].