P14arf Expression Increases Dihydrofolate Reductase Degradation and Paradoxically Results in Resistance to Folate Antagonists in Cells with Nonfunctional P53
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

DNA Damage Checkpoint Dynamics Drive Cell Cycle Phase Transitions
bioRxiv preprint doi: https://doi.org/10.1101/137307; this version posted August 4, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. DNA damage checkpoint dynamics drive cell cycle phase transitions Hui Xiao Chao1,2, Cere E. Poovey1, Ashley A. Privette1, Gavin D. Grant3,4, Hui Yan Chao1, Jeanette G. Cook3,4, and Jeremy E. Purvis1,2,4,† 1Department of Genetics 2Curriculum for Bioinformatics and Computational Biology 3Department of Biochemistry and Biophysics 4Lineberger Comprehensive Cancer Center University of North Carolina, Chapel Hill 120 Mason Farm Road Chapel Hill, NC 27599-7264 †Corresponding Author: Jeremy Purvis Genetic Medicine Building 5061, CB#7264 120 Mason Farm Road Chapel Hill, NC 27599-7264 [email protected] ABSTRACT DNA damage checkpoints are cellular mechanisms that protect the integrity of the genome during cell cycle progression. In response to genotoxic stress, these checkpoints halt cell cycle progression until the damage is repaired, allowing cells enough time to recover from damage before resuming normal proliferation. Here, we investigate the temporal dynamics of DNA damage checkpoints in individual proliferating cells by observing cell cycle phase transitions following acute DNA damage. We find that in gap phases (G1 and G2), DNA damage triggers an abrupt halt to cell cycle progression in which the duration of arrest correlates with the severity of damage. However, cells that have already progressed beyond a proposed “commitment point” within a given cell cycle phase readily transition to the next phase, revealing a relaxation of checkpoint stringency during later stages of certain cell cycle phases. -

Synergistic Tumor Suppression by Combined Inhibition of Telomerase
Synergistic tumor suppression by combined inhibition PNAS PLUS of telomerase and CDKN1A Romi Guptaa, Yuying Donga, Peter D. Solomona, Hiromi I. Wetterstenb, Christopher J. Chengc,d, JIn-Na Mina,e, Jeremy Hensonf,g, Shaillay Kumar Dograh, Sung H. Hwangi, Bruce D. Hammocki, Lihua J. Zhuj, Roger R. Reddelf,g, W. Mark Saltzmanc, Robert H. Weissb,k, Sandy Changa,e, Michael R. Greenl,1, and Narendra Wajapeyeea,1 Departments of aPathology and eLaboratory Medicine, Yale University School of Medicine, New Haven, CT 06510; iDepartment of Entomology and bDivision of Nephrology, Department of Internal Medicine, University of California, Davis, California 95616; Departments of cBiomedical Engineering and dMolecular Biophysics and Biochemistry, Yale University, New Haven, CT 06511; fSydney Medical School, University of Sydney, NSW 2006, Australia; gCancer Research Unit, Children’s Medical Research Institute, Westmead, NSW 2145, Australia; hSingapore Institute of Clinical Sciences, Agency for Science Technology and Research (A*STAR), Brenner Center for Molecular Medicine, Singapore 117609; lHoward Hughes Medical Institute and jPrograms in Gene Function and Expression and Molecular Medicine, University of Massachusetts Medical School, Massachusetts 01605; and kDepartment of Medicine, Mather VA Medical Center, Sacramento, CA 9565 Contributed by Michael R. Green, June 19, 2014 (sent for review June 8, 2014) Tumor suppressor p53 plays an important role in mediating growth dition to its role in cell cycle regulation, p21 has been shown in inhibition upon telomere dysfunction. Here, we show that loss of a variety of studies to repress apoptosis (9–13). the p53 target gene cyclin-dependent kinase inhibitor 1A (CDKN1A, Here,westudytheroleofp21inthe context of telomerase in- also known as p21WAF1/CIP1) increases apoptosis induction following hibition. -

Full Text (PDF)
ResearchArticle DNA Damage–Dependent Translocation of B23 and p19ARF Is Regulated by the Jun N-Terminal Kinase Pathway Orli Yogev,1 Keren Saadon,1 Shira Anzi,1 Kazushi Inoue,2 and Eitan Shaulian1 1Department of Experimental Medicine and Cancer Research, Hebrew University Medical School, Jerusalem, Israel and 2Departments of Pathology/Cancer Biology, Wake Forest University Health Sciences, Winston-Salem, North Carolina Abstract arrest or apoptosis. However, increased tumor development in The dynamic behavior of the nucleolus plays a role in the triple knockout mice nullizygous for ARF, p53, and Mdm2 showed detection of and response to DNA damage of cells. Two that ARF acts also in a p53-independent manner (11). Some of the nucleolar proteins, p14ARF/p19ARF and B23, were shown to p53-independent activity is attributed to its ability to reduce rRNA translocate out of the nucleolus after exposure of cells to DNA- processing (12, 13) and inhibit oncogene-induced transcription damaging agents. This translocation affects multiple cellular (14, 15). functions, such as DNA repair, proliferation, and survival. In ARF augments p53 activity mainly in response to oncogenic this study, we identify a pathway and scrutinize the mecha- stress. Its expression is up-regulated in response to deregulated nisms leading to the translocation of these proteins after oncogenic activity due to elevated transcription governed by several transcription factors, such as E2F (16), c-myc (17), AP-1 exposure of cells to DNA-damaging agents. We show that redistribution of B23 and p19ARF after the exposure to (18), or by oncogenic Ras (19). A second mechanism regulating ARF activity involves the control of its subcellular localization (3). -

Mitosis Vs. Meiosis
Mitosis vs. Meiosis In order for organisms to continue growing and/or replace cells that are dead or beyond repair, cells must replicate, or make identical copies of themselves. In order to do this and maintain the proper number of chromosomes, the cells of eukaryotes must undergo mitosis to divide up their DNA. The dividing of the DNA ensures that both the “old” cell (parent cell) and the “new” cells (daughter cells) have the same genetic makeup and both will be diploid, or containing the same number of chromosomes as the parent cell. For reproduction of an organism to occur, the original parent cell will undergo Meiosis to create 4 new daughter cells with a slightly different genetic makeup in order to ensure genetic diversity when fertilization occurs. The four daughter cells will be haploid, or containing half the number of chromosomes as the parent cell. The difference between the two processes is that mitosis occurs in non-reproductive cells, or somatic cells, and meiosis occurs in the cells that participate in sexual reproduction, or germ cells. The Somatic Cell Cycle (Mitosis) The somatic cell cycle consists of 3 phases: interphase, m phase, and cytokinesis. 1. Interphase: Interphase is considered the non-dividing phase of the cell cycle. It is not a part of the actual process of mitosis, but it readies the cell for mitosis. It is made up of 3 sub-phases: • G1 Phase: In G1, the cell is growing. In most organisms, the majority of the cell’s life span is spent in G1. • S Phase: In each human somatic cell, there are 23 pairs of chromosomes; one chromosome comes from the mother and one comes from the father. -

P14ARF Inhibits Human Glioblastoma–Induced Angiogenesis by Upregulating the Expression of TIMP3
P14ARF inhibits human glioblastoma–induced angiogenesis by upregulating the expression of TIMP3 Abdessamad Zerrouqi, … , Daniel J. Brat, Erwin G. Van Meir J Clin Invest. 2012;122(4):1283-1295. https://doi.org/10.1172/JCI38596. Research Article Oncology Malignant gliomas are the most common and the most lethal primary brain tumors in adults. Among malignant gliomas, 60%–80% show loss of P14ARF tumor suppressor activity due to somatic alterations of the INK4A/ARF genetic locus. The tumor suppressor activity of P14ARF is in part a result of its ability to prevent the degradation of P53 by binding to and sequestering HDM2. However, the subsequent finding of P14ARF loss in conjunction with TP53 gene loss in some tumors suggests the protein may have other P53-independent tumor suppressor functions. Here, we report what we believe to be a novel tumor suppressor function for P14ARF as an inhibitor of tumor-induced angiogenesis. We found that P14ARF mediates antiangiogenic effects by upregulating expression of tissue inhibitor of metalloproteinase–3 (TIMP3) in a P53-independent fashion. Mechanistically, this regulation occurred at the gene transcription level and was controlled by HDM2-SP1 interplay, where P14ARF relieved a dominant negative interaction of HDM2 with SP1. P14ARF-induced expression of TIMP3 inhibited endothelial cell migration and vessel formation in response to angiogenic stimuli produced by cancer cells. The discovery of this angiogenesis regulatory pathway may provide new insights into P53-independent P14ARF tumor-suppressive mechanisms that have implications for the development of novel therapies directed at tumors and other diseases characterized by vascular pathology. Find the latest version: https://jci.me/38596/pdf Research article P14ARF inhibits human glioblastoma–induced angiogenesis by upregulating the expression of TIMP3 Abdessamad Zerrouqi,1 Beata Pyrzynska,1,2 Maria Febbraio,3 Daniel J. -

Loss of P21 Disrupts P14arf-Induced G1 Cell Cycle Arrest but Augments P14arf-Induced Apoptosis in Human Carcinoma Cells
Oncogene (2005) 24, 4114–4128 & 2005 Nature Publishing Group All rights reserved 0950-9232/05 $30.00 www.nature.com/onc Loss of p21 disrupts p14ARF-induced G1 cell cycle arrest but augments p14ARF-induced apoptosis in human carcinoma cells Philipp G Hemmati1,3, Guillaume Normand1,3, Berlinda Verdoodt1, Clarissa von Haefen1, Anne Hasenja¨ ger1, DilekGu¨ ner1, Jana Wendt1, Bernd Do¨ rken1,2 and Peter T Daniel*,1,2 1Department of Hematology, Oncology and Tumor Immunology, University Medical Center Charite´, Campus Berlin-Buch, Berlin-Buch, Germany; 2Max-Delbru¨ck-Center for Molecular Medicine, Berlin-Buch, Germany The human INK4a locus encodes two structurally p16INK4a and p14ARF (termed p19ARF in the mouse), latter unrelated tumor suppressor proteins, p16INK4a and p14ARF of which is transcribed in an Alternative Reading Frame (p19ARF in the mouse), which are frequently inactivated in from a separate exon 1b (Duro et al., 1995; Mao et al., human cancer. Both the proapoptotic and cell cycle- 1995; Quelle et al., 1995; Stone et al., 1995). P14ARF is regulatory functions of p14ARF were initially proposed to usually expressed at low levels, but rapid upregulation be strictly dependent on a functional p53/mdm-2 tumor of p14ARF is triggered by various stimuli, that is, suppressor pathway. However, a number of recent reports the expression of cellular or viral oncogenes including have implicated p53-independent mechanisms in the E2F-1, E1A, c-myc, ras, and v-abl (de Stanchina et al., regulation of cell cycle arrest and apoptosis induction by 1998; Palmero et al., 1998; Radfar et al., 1998; Zindy p14ARF. Here, we show that the G1 cell cycle arrest et al., 1998). -

Alterations of P14arf, P53, and P73 Genes Involved in the E2F-1-Mediated Apoptotic Pathways in Non-Small Cell Lung Carcinoma
[CANCER RESEARCH 61, 5636–5643, July 15, 2001] Alterations of p14ARF, p53, and p73 Genes Involved in the E2F-1-mediated Apoptotic Pathways in Non-Small Cell Lung Carcinoma Siobhan A. Nicholson,1 Nader T. Okby,1 Mohammed A. Khan, Judith A. Welsh, Mary G. McMenamin, William D. Travis, James R. Jett, Henry D. Tazelaar, Victor Trastek, Peter C. Pairolero, Paul G. Corn, James G. Herman, Lance A. Liotta, Neil E. Caporaso, and Curtis C. Harris2 Laboratory of Human Carcinogenesis, National Cancer Institute, Bethesda, Maryland 20892 [S. A. N., M. A. K., J. A. W., M. G. M., L. A. L., N. E. C., C. C. H.]; Orange Pathology Associates, Middleton, New York 10940 [N. T. O.]; Armed Forces Institute of Pathology, Washington, DC 20306 [S. A. N., W. D. T.]; Mayo Clinic, Rochester, Minnesota 55905 [J. R. J., H. D. T., V. T., P. C. P.]; and The Johns Hopkins Oncology Center, Baltimore, Maryland 21231 [P. G. C., J. G. H.] ABSTRACT encoded by a separate exon 1 that lies ϳ20 kb upstream of exon 1␣ and shares exons 2 and 3 as read in an ARF, giving rise to a protein ARF Overexpression of E2F-1 induces apoptosis by both a p14 -p53- and INK4a ARF completely unrelated to p16 (14). Despite its unrelated structure, a p73-mediated pathway. p14 is the alternate tumor suppressor prod- ARF p14 also is capable of causing cell cycle arrest in G1 and G2. uct of the INK4a/ARF locus that is inactivated frequently in lung carci- ARF nogenesis. Because p14ARF stabilizes p53, it has been proposed that the loss p14 binds to and antagonizes the actions of MDM2, a negative of p14ARF is functionally equivalent to a p53 mutation. -

The Cell Cycle Coloring Worksheet
Name: Date: Period: The Cell Cycle Coloring Worksheet Label the diagram below with the following labels: Anaphase Interphase Mitosis Cell division (M Phase) Interphase Prophase Cytokinesis Interphase S-DNA replication G1 – cell grows Metaphase Telophase G2 – prepares for mitosis Then on the diagram, lightly color the G1 phase BLUE, the S phase YELLOW, the G2 phase RED, and the stages of mitosis ORANGE. Color the arrows indicating all of the interphases in GREEN. Color the part of the arrow indicating mitosis PURPLE and the part of the arrow indicating cytokinesis YELLOW. M-PHASE YELLOW: GREEN: CYTOKINESIS INTERPHASE PURPLE: TELOPHASE MITOSIS ANAPHASE ORANGE METAPHASE BLUE: G1: GROWS PROPHASE PURPLE MITOSIS RED:G2: PREPARES GREEN: FOR MITOSIS INTERPHASE YELLOW: S PHASE: DNA REPLICATION GREEN: INTERPHASE Use the diagram and your notes to answer the following questions. 1. What is a series of events that cells go through as they grow and divide? CELL CYCLE 2. What is the longest stage of the cell cycle called? INTERPHASE 3. During what stage does the G1, S, and G2 phases happen? INTERPHASE 4. During what phase of the cell cycle does mitosis and cytokinesis occur? M-PHASE 5. During what phase of the cell cycle does cell division occur? MITOSIS 6. During what phase of the cell cycle is DNA replicated? S-PHASE 7. During what phase of the cell cycle does the cell grow? G1,G2 8. During what phase of the cell cycle does the cell prepare for mitosis? G2 9. How many stages are there in mitosis? 4 10. Put the following stages of mitosis in order: anaphase, prophase, metaphase, and telophase. -

Working on Genomic Stability: from the S-Phase to Mitosis
G C A T T A C G G C A T genes Review Working on Genomic Stability: From the S-Phase to Mitosis Sara Ovejero 1,2,3,* , Avelino Bueno 1,4 and María P. Sacristán 1,4,* 1 Instituto de Biología Molecular y Celular del Cáncer (IBMCC), Universidad de Salamanca-CSIC, Campus Miguel de Unamuno, 37007 Salamanca, Spain; [email protected] 2 Institute of Human Genetics, CNRS, University of Montpellier, 34000 Montpellier, France 3 Department of Biological Hematology, CHU Montpellier, 34295 Montpellier, France 4 Departamento de Microbiología y Genética, Universidad de Salamanca, Campus Miguel de Unamuno, 37007 Salamanca, Spain * Correspondence: [email protected] (S.O.); [email protected] (M.P.S.); Tel.: +34-923-294808 (M.P.S.) Received: 31 January 2020; Accepted: 18 February 2020; Published: 20 February 2020 Abstract: Fidelity in chromosome duplication and segregation is indispensable for maintaining genomic stability and the perpetuation of life. Challenges to genome integrity jeopardize cell survival and are at the root of different types of pathologies, such as cancer. The following three main sources of genomic instability exist: DNA damage, replicative stress, and chromosome segregation defects. In response to these challenges, eukaryotic cells have evolved control mechanisms, also known as checkpoint systems, which sense under-replicated or damaged DNA and activate specialized DNA repair machineries. Cells make use of these checkpoints throughout interphase to shield genome integrity before mitosis. Later on, when the cells enter into mitosis, the spindle assembly checkpoint (SAC) is activated and remains active until the chromosomes are properly attached to the spindle apparatus to ensure an equal segregation among daughter cells. -

S-Phase Entry, Activation of E2F-Responsive Genes, and Apoptosis
Proc. Natl. Acad. Sci. USA Vol. 92, pp. 5436-5440, June 1995 Medical Sciences Deficiency of retinoblastoma protein leads to inappropriate S-phase entry, activation of E2F-responsive genes, and apoptosis (tumor supressor gene/cell cycle control/programmed cell death) ALEXANDRU ALMASAN*tt, YUXIN YIN*t§, RUTH E. KELLY*, EVA Y.-H. P. LEES, ALLAN BRADLEYII, WEIWEI LI**, JOSEPH R. BERTINO**, AND GEOFFREY M. WAHL*tt *Gene Expression Laboratory, The Salk Institute, 10010 North Torrey Pines Road, La Jolla, CA 92037; tCenter for Molecular Medicine and Institute of Biotechnology, The University of Texas Health Science Center at San Antonio, San Antonio, TX 78284; I'Department of Molecular and Human Genetics and Howard Hughes Medical Institute, Baylor College of Medicine, Houston, TX 77030; and **Program for Molecular Pharmacology and Therapeutics, Memorial Sloan-Kettering Cancer Center, New York, NY 10021 Communicated by Leslie E. Orgel, The Salk Institute for Biological Studies, San Diego, CA, January 24, 1995 (received for review December 12, 1994) ABSTRACT The retinoblastoma susceptibility gene (Rb) thymidylate synthase (TS) and other genes necessary for entry participates in controlling the G1/S-phase transition, pre- into and progression through S phase (10, 14). sumably by binding and inactivating E2F transcription acti- The data obtained from studies with viral oncoproteins vator family members. Mouse embryonic fibroblasts (MEFs) suggest that Rb protein acts by sequestering essential factors with no, one, or two inactivated Rb genes were used to determine required for G1/S progression. However, each of these onco- the specific contributions of Rb protein to cell cycle progres- proteins binds other cellular factors such as the Rb-related sion and gene expression. -

Expression of P16 INK4A and P14 ARF in Hematological Malignancies
Leukemia (1999) 13, 1760–1769 1999 Stockton Press All rights reserved 0887-6924/99 $15.00 http://www.stockton-press.co.uk/leu Expression of p16INK4A and p14ARF in hematological malignancies T Taniguchi1, N Chikatsu1, S Takahashi2, A Fujita3, K Uchimaru4, S Asano5, T Fujita1 and T Motokura1 1Fourth Department of Internal Medicine, University of Tokyo, School of Medicine; 2Division of Clinical Oncology, Cancer Chemotherapy Center, Cancer Institute Hospital; 3Department of Hematology, Showa General Hospital; 4Third Department of Internal Medicine, Teikyo University, School of Medicine; and 5Department of Hematology/Oncology, Institute of Medical Science, University of Tokyo, Tokyo, Japan The INK4A/ARF locus yields two tumor suppressors, p16INK4A tumor suppressor genes.10,11 In human hematological malig- ARF and p14 , and is frequently deleted in human tumors. We nancies, their inactivation occurs mainly by means of homo- studied their mRNA expressions in 41 hematopoietic cell lines and in 137 patients with hematological malignancies; we used zygous deletion or promoter region hypermethylation a quantitative reverse transcription-PCR assay. Normal periph- (reviewed in Ref. 12). In tumors such as pancreatic adenocar- eral bloods, bone marrow and lymph nodes expressed little or cinomas, esophageal squamous cell carcinomas and familial undetectable p16INK4A and p14ARF mRNAs, which were readily melanomas, p16INK4A is often inactivated by point mutation, detected in 12 and 17 of 41 cell lines, respectively. Patients with 12 INK4A which is not the case in hematological malignancies. On hematological malignancies frequently lacked p16 INK4C INK4D ARF the other hand, genetic aberrations of p18 or p19 expression (60/137) and lost p14 expression less frequently 12 (19/137, 13.9%). -
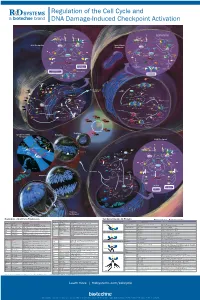
Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation
RnDSy-lu-2945 Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation IR UV IR Stalled Replication Forks/ BRCA1 Rad50 Long Stretches of ss-DNA Rad50 Mre11 BRCA1 Nbs1 Rad9-Rad1-Hus1 Mre11 RPA MDC1 γ-H2AX DNA Pol α/Primase RFC2-5 MDC1 Nbs1 53BP1 MCM2-7 53BP1 γ-H2AX Rad17 Claspin MCM10 Rad9-Rad1-Hus1 TopBP1 CDC45 G1/S Checkpoint Intra-S-Phase RFC2-5 ATM ATR TopBP1 Rad17 ATRIP ATM Checkpoint Claspin Chk2 Chk1 Chk2 Chk1 ATR Rad50 ATRIP Mre11 FANCD2 Ubiquitin MDM2 MDM2 Nbs1 CDC25A Rad50 Mre11 BRCA1 Ub-mediated Phosphatase p53 CDC25A Ubiquitin p53 FANCD2 Phosphatase Degradation Nbs1 p53 p53 CDK2 p21 p21 BRCA1 Ub-mediated SMC1 Degradation Cyclin E/A SMC1 CDK2 Slow S Phase CDC45 Progression p21 DNA Pol α/Primase Slow S Phase p21 Cyclin E Progression Maintenance of Inhibition of New CDC6 CDT1 CDC45 G1/S Arrest Origin Firing ORC MCM2-7 MCM2-7 Recovery of Stalled Replication Forks Inhibition of MCM10 MCM10 Replication Origin Firing DNA Pol α/Primase ORI CDC6 CDT1 MCM2-7 ORC S Phase Delay MCM2-7 MCM10 MCM10 ORI Geminin EGF EGF R GAB-1 CDC6 CDT1 ORC MCM2-7 PI 3-Kinase p70 S6K MCM2-7 S6 Protein Translation Pre-RC (G1) GAB-2 MCM10 GSK-3 TSC1/2 MCM10 ORI PIP2 TOR Promotes Replication CAK EGF Origin Firing Origin PIP3 Activation CDK2 EGF R Akt CDC25A PDK-1 Phosphatase Cyclin E/A SHIP CIP/KIP (p21, p27, p57) (Active) PLCγ PP2A (Active) PTEN CDC45 PIP2 CAK Unwinding RPA CDC7 CDK2 IP3 DAG (Active) Positive DBF4 α Feedback CDC25A DNA Pol /Primase Cyclin E Loop Phosphatase PKC ORC RAS CDK4/6 CDK2 (Active) Cyclin E MCM10 CDC45 RPA IP Receptor