Weigelaspecies and Cultivar Genome Size and Ploidy Estimations: Shrub Breeding©
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Phylogeny and Phylogenetic Taxonomy of Dipsacales, with Special Reference to Sinadoxa and Tetradoxa (Adoxaceae)
PHYLOGENY AND PHYLOGENETIC TAXONOMY OF DIPSACALES, WITH SPECIAL REFERENCE TO SINADOXA AND TETRADOXA (ADOXACEAE) MICHAEL J. DONOGHUE,1 TORSTEN ERIKSSON,2 PATRICK A. REEVES,3 AND RICHARD G. OLMSTEAD 3 Abstract. To further clarify phylogenetic relationships within Dipsacales,we analyzed new and previously pub- lished rbcL sequences, alone and in combination with morphological data. We also examined relationships within Adoxaceae using rbcL and nuclear ribosomal internal transcribed spacer (ITS) sequences. We conclude from these analyses that Dipsacales comprise two major lineages:Adoxaceae and Caprifoliaceae (sensu Judd et al.,1994), which both contain elements of traditional Caprifoliaceae.Within Adoxaceae, the following relation- ships are strongly supported: (Viburnum (Sambucus (Sinadoxa (Tetradoxa, Adoxa)))). Combined analyses of C ap ri foliaceae yield the fo l l ow i n g : ( C ap ri folieae (Diervilleae (Linnaeeae (Morinaceae (Dipsacaceae (Triplostegia,Valerianaceae)))))). On the basis of these results we provide phylogenetic definitions for the names of several major clades. Within Adoxaceae, Adoxina refers to the clade including Sinadoxa, Tetradoxa, and Adoxa.This lineage is marked by herbaceous habit, reduction in the number of perianth parts,nectaries of mul- ticellular hairs on the perianth,and bifid stamens. The clade including Morinaceae,Valerianaceae, Triplostegia, and Dipsacaceae is here named Valerina. Probable synapomorphies include herbaceousness,presence of an epi- calyx (lost or modified in Valerianaceae), reduced endosperm,and distinctive chemistry, including production of monoterpenoids. The clade containing Valerina plus Linnaeeae we name Linnina. This lineage is distinguished by reduction to four (or fewer) stamens, by abortion of two of the three carpels,and possibly by supernumerary inflorescences bracts. Keywords: Adoxaceae, Caprifoliaceae, Dipsacales, ITS, morphological characters, phylogeny, phylogenetic taxonomy, phylogenetic nomenclature, rbcL, Sinadoxa, Tetradoxa. -

Maryum Bhatti1, Aaron Lee1, Hiba Rahman-Vyas1, Dianella G
NAC SUBFAMILY 1A GENE TREE Combining benchwork and bioinformatics to reconstruct the evolutionary history of CUP- SHAPED COTYLEDON in honeysuckles and relatives Maryum Bhatti1, Aaron Lee1, Hiba Rahman-Vyas1, Dianella G. Howarth2, Michael J. Donoghue3, Wendy L. Clement1 1Department of Biology, The College of New Jersey, Ewing, NJ; 2Department of Biological Sciences, St. John’s University, Jamaica, NY; 3Department of Ecology and Evolution, Yale University, New Haven, CT RECOVERING CUC FROM INTRODUCTION DIPSACALES Context Fusion among adjacent parts is a phenomena that occurs Sampling and Isolating CUC throughout flowering plants. CUP-SHAPED COTYLEDON, CUC, • 38 species were selected across the Dipsacales for a member of the NAC Subfamily 1a transcription factors, and has been shown to affect organ boundary formation.1,2 Honeysuckles, species that had no available genomic resources or Lonicera (Caprifoliaceae, Dipsacales), are known for fusing • Degenerate primers were designed to isolate exons petals into long tubes and also exhibit fusion among ovaries, bracts 1 and 2 of CUC1/2 and CUC3 based on reference and leaves. Variation in fusion across 160 species of Lonicera genomes (e.g., Arabidopsis, Petunia, Snapdragon) make them an excellent system to investigate the evolution of • CUC specific primers were also created including fusion. all introns and exons of CUC Goals • Successful reactions near 500 bp were isolated and • Recover CUC from the phylogenetic diversity of Dipsacales, cloned using PCR and cloning; sample target species in which no • Multiple clones per species were sequenced at the genomic data is available. Yale Sequencing on the Hill Facility • Reconstruct a gene tree for NAC Subfamily 1a and CUC using • Sequences were assembled and manually edited both direct sequences gained in this study and data extracted 3 from available genomic resources. -

Old Fashioned Weigela (Caprifoliaceae) ------Weigela Florida Is Valued for Its Use As a Specimen Or Assets Mixed Border Plant
Weigela florida - Old Fashioned Weigela (Caprifoliaceae) ----------------------------------------------------------------------------------------------- Weigela florida is valued for its use as a specimen or Assets mixed border plant. Plant Old Fashioned Weigela -profusion of colorful flowers around windows so its abundant pink flowers can be -relatively low maintenance admired each spring. -adaptable and urban tolerant Liabilities FEATURES -becomes scraggly, unkempt, and larger than Form intended unless properly pruned and managed -deciduous, spreading, dense, rounded shrub reaching Habitat 6-9' tall x 9-12' wide, dense, arching, eventually to Zones 4 to 8 the ground Native to Japan -moderate to fast growth rate; low maintenance. Culture SELECTIONS -Weigela grows best in a sunny location and moist Alternates soil. Shaded plants are straggly. Weigela also -other spring-flowering shrubs (Viburnum, dislikes crowding. Rhododendron, Deutzia gracilis, etc.) -transplants well Cultivars - Variants - Related species -some annual dieback and winter injury make annual -many cultivars (at least 170), including variegated pruning necessary. and dwarfs: -becomes rangy unless pruned, no serious pests or -'Bristol Ruby' - red flowers produced sporadically diseases. throughout the growing season. Foliage -'Bristol Snowflake' - white flowers flushed with -leaves simple, opposite, elliptic, 2-4" long x 1" wide, pink. acuminate (pointed tip), rounded at base, serrate, -'Carnaval' - red, white and pink flowers are produced short petiole. on the same plant. -leaf color is medium to dark green in summer, -'Evita' - a slow growing, compact plant with bright turning brownish in autumn. red flowers produced in spring and then periodically. Flowers -'Java Red' - a hardy plant with purplish foliage and deep pink flowers. -'Minuet' - a hardy cultivar with purplish foliage and flowers that are red, purple and yellow. -

IHCA Recommended Plant List
Residential Architectural Review Committee Recommended Plant List Plant Materials The following plant materials are intended to guide tree and shrub ADDITIONS to residential landscapes at Issaquah Highlands. Lot sizes, shade, wind and other factors place size and growth constraints on plants, especially trees, which are suitable for addition to existing landscapes. Other plant materials may be considered that have these characteristics and similar maintenance requirements. Additional species and varieties may be selected if authorized by the Issaquah Highlands Architectural Review Committee. This list is not exhaustive but does cover most of the “good doers” for Issaquah Highlands. Our microclimate is colder and harsher than those closer to Puget Sound. Plants not listed should be used with caution if their performance has not been observed at Issaquah Highlands. * Drought-tolerant plant ** Requires well-drained soil DECIDUOUS TREES: Small • Acer circinatum – Vine Maple • Acer griseum – Paperbark Maple • *Acer ginnala – Amur Maple • Oxydendrum arboreum – Sourwood • Acer palmation – Japanese Maple • *Prunus cerasifera var. – Purple Leaf Plum varieties • Amelanchier var. – Serviceberry varieties • Styrax japonicus – Japanese Snowbell • Cornus species, esp. kousa Medium • Acer rufinerve – Redvein Maple • Cornus florida (flowering dogwood) • *Acer pseudoplatanus – Sycamore Maple • Acer palmatum (Japanese maple, many) • • *Carpinus betulus – European Hornbeam Stewartia species (several) • *Parrotia persica – Persian Parrotia Columnar Narrow -

Shrubs of the Year
GrowingShrubs & Landscape of the Guide Year LOW SCAPE® AT LAST® FIRE LIGHT® SONIC BLOOM® Mound Aronia Rosa Hydrangea Weigela series Aronia melanocarpa LOW SCAPE ® Mound Aronia melanocarpa ‘UCONNAM165’, pp#28,789, cbraf Durable native groundcover shrub Looking for a replacement for Rhus ‘Gro-Low’? Check out this remarkable new dwarf Aronia. White spring • SIZE & SHAPE 1-2’ TALL flowers are just the start of the show: glossy green foliage turns brilliant red in fall, and black fruit are 2’ WIDE quite ornamental. Quite possibly the perfect landscape plant, it’s adaptable to most soils, doesn’t need 10’ pruning, and provides multi-season interest in the landscape! One just isn’t enough, so use it for mass plantings and edging. NATIVE | DEER-RESISTANT | FALL COLOR | DWARF • SPACING 2-3’ NATURALIZING | RAIN GARDENS | SOIL STABILIZATION • USDA 3, AHS 9 MAY BE TRIMMED AFTER FLOWERING • FULL SUN PART SHADE www.provenwinners-shrubs.com LOW SCAPE® MOUND Aronia melanocarpa 'UCONNAM165' pp#28,789, cbraf Landscape Info: Features & Benefits: Common name: chokeberry - Native to North America USDA/AHS zones: USDA 3/AHS 9 - Tolerant of a wide range of soil, light, and moisture conditions; deer resistant Mature height: 3-5’/.9-1.5 m - Much smaller than conventional aronias and Part shade-full suited to a variety of landscape applications Exposure: sun - White flowers in spring, red-orange fall color, black fruits in late summer attract birds Irrigation: Average Cultural info: Grower Tips: pH Level: Slightly acidic: 5.6 - 6.2 Needs less trimming than other aronias (including Low Scape Nutrient Needs: Average Hedger) to attain an attractive habit; trim much like you would boxwood. -

A Check-List of Cultivar Names in Weigela
ARNOLDIA A continuation of the BULLETIN OF POPULAR INFORMATION of the Arnold Arboretum, Harvard University VOLUME 25 DECEMBER 17, 1965 NUMBERS 9-111 A CHECK-LIST OF CULTIVAR NAMES IN WEIGELA IN 1929 L. H. BAILEY CONSIDERED "The Case of Diervilla and Weigela" (Gent. Herb. 2: 39-54. 1929) and concluded that the two genera were truly distinct morphologically and geographically, as well as horticulturally. His review of the history of these two genera is complete and need not be repeated. A third taxon at the generic level, Calyptrostigma, was also recognized by Bailey as containing "at least two species in Eastern Asia..." and "little known in cultivation, as they are usually rather difficult to grow." Nakai (Jour. Jap. Bot. 12: 1-17. 1936) did not agree with Bailey and combined Weigela and Diervilla and proposed two new genera. He published the genus Weigelastrum to contain Diervilla maximowiczii and he substituted the name Macrodiervilla for the illegitimate Calyptrostigma which Bailey had accepted under different rules of nomenclature. Rehder ( Jour. Arnold Arb. 20: 429-431. 1939) reduced both of Nakai’s genera to sectional rank and accepted Bailey’s recognition of Weigela and Diervilla. Rehder’s treat- ment is followed by most American horticulturists, although some nurserymen still list species and varieties of Weigela under the name Diervilla, or under both names, and often indiscriminately. The situation is confused in modern treatments from Europe. In England, Bean’s Trees and Shrubs Hardy in the British Isles recognizes only Diervilla and this is also the treatment given in the Royal Horticultural Society’s Dictionary off Gardening. -

Phylogenetics of the Caprifolieae and Lonicera (Dipsacales)
Systematic Botany (2008), 33(4): pp. 776–783 © Copyright 2008 by the American Society of Plant Taxonomists Phylogenetics of the Caprifolieae and Lonicera (Dipsacales) Based on Nuclear and Chloroplast DNA Sequences Nina Theis,1,3,4 Michael J. Donoghue,2 and Jianhua Li1,4 1Arnold Arboretum of Harvard University, 22 Divinity Ave, Cambridge, Massachusetts 02138 U.S.A. 2Department of Ecology and Evolutionary Biology, Yale University, P.O. Box 208106, New Haven, Conneticut 06520-8106 U.S.A. 3Current Address: Plant Soil, and Insect Sciences, University of Massachusetts at Amherst, Amherst, Massachusetts 01003 U.S.A. 4Authors for correspondence ([email protected]; [email protected]) Communicating Editor: Lena Struwe Abstract—Recent phylogenetic analyses of the Dipsacales strongly support a Caprifolieae clade within Caprifoliaceae including Leycesteria, Triosteum, Symphoricarpos, and Lonicera. Relationships within Caprifolieae, however, remain quite uncertain, and the monophyly of Lonicera, the most species-rich of the traditional genera, and its subdivisions, need to be evaluated. In this study we used sequences of the ITS region of nuclear ribosomal DNA and five chloroplast non-coding regions (rpoB–trnC spacer, atpB–rbcL spacer, trnS–trnG spacer, petN–psbM spacer, and psbM–trnD spacer) to address these problems. Our results indicate that Heptacodium is sister to Caprifolieae, Triosteum is sister to the remaining genera within the tribe, and Leycesteria and Symphoricarpos form a clade that is sister to a monophyletic Lonicera. Within Lonicera, the major split is between subgenus Caprifolium and subgenus Lonicera. Within subgenus Lonicera, sections Coeloxylosteum, Isoxylosteum, and Nintooa are nested within the paraphyletic section Isika. Section Nintooa may also be non-monophyletic. -

Phylogeny and Biogeography of Valerianaceae (Dipsacales) with Special Reference to the South American Valerians Charles D.Bell Ã,1, Michael J.Donoghue
ARTICLE IN PRESS Organisms, Diversity & Evolution 5 (2005) 147–159 www.elsevier.de/ode Phylogeny and biogeography of Valerianaceae (Dipsacales) with special reference to the South American valerians Charles D.Bell Ã,1, Michael J.Donoghue Department of Ecology and Evolutionary Biology, Yale University, New Haven, CT 065211, USA Received 22 April 2004; accepted 26 October 2004 Abstract Species of Valerianaceae are a common component of the alpine flora throughout the Northern Hemisphere as well as the Andes of South America.Sequence data from three chloroplast markers ( psbA-trnH intron, trnK-matK intron, and the trnL-F region) along with the internal transcribed spacer region (ITS) of nuclear ribosomal DNA were used to infer relationships within Valerianaceae.Both genomes, as well as a combined data set, provide support for the major clades within the group and do not support a monophyletic Valeriana.In addition, these data indicate that Plectritis is nested within South American Valeriana, as opposed to being sister to Centhranthus as previously hypothesized. Valerianaceae appear to have originated in Asia, probably in the Himalayas, and subsequently to have dispersed several times to Europe and to the New World.Our results imply that Valerianaceae colonized South America on multiple occasions from the north.In one of these cases there appears to have been a substantial and rapid radiation, primarily in the high elevation paramo habitat.A variety of methods were used to estimate divergence times to determine when Valerianaceae might have colonized South America.Regardless of the method and fossil constraints applied, our estimates suggest that Valerianaceae colonized South America prior to the formation of the Isthmus of Panama. -

Dipsacales Phylogeny Based on Chloroplast Dna Sequences
DIPSACALES PHYLOGENY BASED ON CHLOROPLAST DNA SEQUENCES CHARLES D. BELL,1, 2 ERIKA J. EDWARDS,1 SANG-TAE KIM,1 AND MICHAEL J. DONOGHUE 1 Abstract. Eight new rbcL DNA sequences and 15 new sequences from the 5' end of the chloroplast ndhF gene were obtained from representative Dipsacales and outgroup taxa. These were analyzed in combination with pre- viously published sequences for both regions. In addition, sequence data from the entire ndhF gene, the trnL-F intergenic spacer region,the trnL intron,the matK region, and the rbcL-atpB intergenic spacer region were col- lected for 30 taxa within Dipsacales. Phylogenetic relationships were inferred using maximum parsimony and maximum likelihood methods. Inferred tree topologies are in strong agreement with previous results from sep- arate and combined analyses of rbcL and morpholo gy, and confidence in most major clades is now very high. Concerning controversial issues, we conclude that Dipsacales in the traditional sense is a monophyletic group and that Triplostegia is more closely related to Dipsacaceae than it is to Valerianaceae. Heptacodium is only weakly supported as the sister group of the Caprifolieae (within which relationships remain largely unresolved), and the exact position of Diervilleae is uncertain. Within Morinaceae, Acanthocalyx is the sister group of Morina plus Cryptothladia. Dipsacales now provides excellent opportunities for comparative studies, but it will be important to check the congruence of chloroplast results with those based on data from other genomes. Keywords: Dipsacales, Adoxaceae, Caprifoliaceae, Morinaceae, Dipsacaceae, Valerianaceae, phylogeny, chloroplast DNA. The Dipsacales has traditionally included the Linnaea). It excludes Adoxa and its relatives, as C ap ri foliaceae (s e n s u l at o, i . -
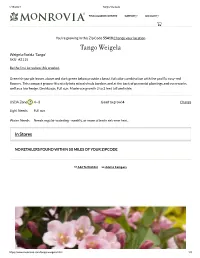
Tango Weigela
1/15/2021 Tango Weigela FIND A GARDEN CENTER SUPPORT ACCOUNT You're growing in this Zip Code 55410 Change your location Tango Weigela Weigela orida 'Tango' SKU #2115 Be the rst to review this product Greenish-purple leaves above and dark green below provide a beautiful color combination with the prolic rosy-red owers. This compact grower ts nicely into mixed shrub borders and at the back of perennial plantings and even works well as a low hedge. Deciduous. Full sun. Moderate growth 2 to 3 feet tall and wide. USDA Zone ? 4 - 8 Good to grow!4 Change Light Needs Full sun Water Needs Needs regular watering - weekly, or more often in extreme heat. In Stores NO RETAILERS FOUND WITHIN 50 MILES OF YOUR ZIPCODE Add To Wishlist Add to Compare https://www.monrovia.com/tango-weigela.html 1/3 1/15/2021 Tango Weigela DETAIL Botanical Pronunciation wy-GEE-la FLOR-ih-duh Average Size at Maturity Compact shrub 2 to 3 ft. tall and wide. Bloom Time Summer Deciduous/ Evergreen Deciduous Flower Attribute Long Bloom Season, Showy Flowers Flower Color Red Foliage Color Green Garden Style Cottage, Rustic Growth Habit Compact Growth Rate Moderate Landscape Use Firescaping/Fire Wise Light Needs Full sun https://www.monrovia.com/tango-weigela.html 2/3 1/15/2021 Tango Weigela Soil Needs All-Purpose Plant Food Special Feature Attracts Butteries, Extreme Cold Hardiness, Compact Form Water Needs Moderate Watering Needs Needs regular watering - weekly, or more often in extreme heat. Key Feature Summer Flowering CARE REVIEWS This Plant's Growing Zones: 4 - 8 Your USDA Cold Hardiness Zone: 4 Good to grow! Change Zip Code https://www.monrovia.com/tango-weigela.html 3/3. -

Plants for Long-Season Bloom
Visit us on the Web: www.gardeninghelp.org Plants for Long-season Bloom The following plants were selected from plants that have displayed a long-season of bloom at the Missouri Botanical Garden based on several years of bloom data. Grasses that hold their flower stalk well into winter are also included. In many cases, only the species or a good, representative cultivar of a species has been included. For example, many cultivars of purple coneflower, Echinacea purpurea are good, long-season bloomers, but only the species has been listed. Grasses* Andropogon gerardii big blue stem Bouteloua curtipendula sideoats grass Calamagrostis x acutiflora 'Karl Foerster' feather reed grass Chasmanthium latifolium northern sea oats Elymus canadensis Canada wild rye Eragrostis trichodes sand lovegrass Hakonechloa macra 'Aureola' Japanese forest grass Miscanthus sinensis 'Morning Light' eulalia Panicum virgatum 'Dallas Blues' switch grass Pennisetum alopecuroides 'Hameln' fountain grass Saccharum ravennae plume grass Sporobolus heterolepis prairie dropseed * have persistent flower stalk into winter for long-season appeal Perennials – Full Sun Achillea 'Coronation Gold' yarrow Asclepias tuberosa butterfly weed Aster novae-angliae 'Purple Dome' New England aster Aster oblongifolius aromatic aster Boltonia asteroides var. latisquama 'Snowbank' false aster Callirhoe involucrata purple poppy mallow Dianthus 'Feuerhexe' FIREWITCH cheddar pink Eryngium yuccifolium rattlesnake master Gaura lindheimeri gaura Glandularia canadensis rose verbena Goniolimon tataricum -

International Union for the Protection of New Varieties of Plants
E TG/148/3(proj.1) ORIGINAL: English DATE: 2021-04-28 INTERNATIONAL UNION FOR THE PROTECTION OF NEW VARIETIES OF PLANTS Geneva DRAFT * WEIGELA UPOV Code(s): WEIGE Weigela Thunb. GUIDELINES FOR THE CONDUCT OF TESTS FOR DISTINCTNESS, UNIFORMITY AND STABILITY prepared by experts from France to be considered by the Technical Working Party for Ornamental Plants and Forest Trees at its fifty-third session, to be held in Roelofarendsveen, Netherlands, from 2021-06-07 to 2021-06-11 Disclaimer: this document does not represent UPOV policies or guidance Alternative names:* Botanical name English French German Spanish Weigela Thunb. Weigela Weigela Weigela Weigela The purpose of these guidelines (“Test Guidelines”) is to elaborate the principles contained in the General Introduction (document TG/1/3), and its associated TGP documents, into detailed practical guidance for the harmonized examination of distinctness, uniformity and stability (DUS) and, in particular, to identify appropriate characteristics for the examination of DUS and production of harmonized variety descriptions. ASSOCIATED DOCUMENTS These Test Guidelines should be read in conjunction with the General Introduction and its associated TGP documents. * These names were correct at the time of the introduction of these Test Guidelines but may be revised or updated. [Readers are advised to consult the UPOV Code, which can be found on the UPOV Website (www.upov.int), for the latest information.] TG/148/3(proj.1) Weigela, 2021-04-28 2 TABLE OF CONTENTS PAGE 1. SUBJECT OF THESE TEST GUIDELINES.......................................................................................................... 3 2. MATERIAL REQUIRED..................................................................................................................... 3 3. METHOD OF EXAMINATION............................................................................................................... 3 3.1 Number of Growing Cycles................................................................................................................