Starviewer and Its Comparison with Other Open-Source DICOM Viewers Using a Novel Hierarchical Evaluation Framework
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Management of Large Sets of Image Data Capture, Databases, Image Processing, Storage, Visualization Karol Kozak
Management of large sets of image data Capture, Databases, Image Processing, Storage, Visualization Karol Kozak Download free books at Karol Kozak Management of large sets of image data Capture, Databases, Image Processing, Storage, Visualization Download free eBooks at bookboon.com 2 Management of large sets of image data: Capture, Databases, Image Processing, Storage, Visualization 1st edition © 2014 Karol Kozak & bookboon.com ISBN 978-87-403-0726-9 Download free eBooks at bookboon.com 3 Management of large sets of image data Contents Contents 1 Digital image 6 2 History of digital imaging 10 3 Amount of produced images – is it danger? 18 4 Digital image and privacy 20 5 Digital cameras 27 5.1 Methods of image capture 31 6 Image formats 33 7 Image Metadata – data about data 39 8 Interactive visualization (IV) 44 9 Basic of image processing 49 Download free eBooks at bookboon.com 4 Click on the ad to read more Management of large sets of image data Contents 10 Image Processing software 62 11 Image management and image databases 79 12 Operating system (os) and images 97 13 Graphics processing unit (GPU) 100 14 Storage and archive 101 15 Images in different disciplines 109 15.1 Microscopy 109 360° 15.2 Medical imaging 114 15.3 Astronomical images 117 15.4 Industrial imaging 360° 118 thinking. 16 Selection of best digital images 120 References: thinking. 124 360° thinking . 360° thinking. Discover the truth at www.deloitte.ca/careers Discover the truth at www.deloitte.ca/careers © Deloitte & Touche LLP and affiliated entities. Discover the truth at www.deloitte.ca/careers © Deloitte & Touche LLP and affiliated entities. -

A 3D Interactive Multi-Object Segmentation Tool Using Local Robust Statistics Driven Active Contours
A 3D interactive multi-object segmentation tool using local robust statistics driven active contours The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Gao, Yi, Ron Kikinis, Sylvain Bouix, Martha Shenton, and Allen Tannenbaum. 2012. A 3D Interactive Multi-Object Segmentation Tool Using Local Robust Statistics Driven Active Contours. Medical Image Analysis 16, no. 6: 1216–1227. doi:10.1016/j.media.2012.06.002. Published Version doi:10.1016/j.media.2012.06.002 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:28548930 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA NIH Public Access Author Manuscript Med Image Anal. Author manuscript; available in PMC 2013 August 01. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Med Image Anal. 2012 August ; 16(6): 1216–1227. doi:10.1016/j.media.2012.06.002. A 3D Interactive Multi-object Segmentation Tool using Local Robust Statistics Driven Active Contours Yi Gaoa,*, Ron Kikinisb, Sylvain Bouixa, Martha Shentona, and Allen Tannenbaumc aPsychiatry Neuroimaging Laboratory, Brigham & Women's Hospital, Harvard Medical School, Boston, MA 02115 bSurgical Planning Laboratory, Brigham & Women's Hospital, Harvard Medical School, Boston, MA 02115 cDepartments of Electrical and Computer Engineering and Biomedical Engineering, Boston University, Boston, MA 02115 Abstract Extracting anatomical and functional significant structures renders one of the important tasks for both the theoretical study of the medical image analysis, and the clinical and practical community. -

Qualitative Comparison of Conventional and Oblique MRI for Detection of Herniated Spinal Discs
Qualitative Comparison of Conventional and Oblique MRI for Detection of Herniated Spinal Discs Doug Dean Final Project Presentation ENGN 2500: Medical Image Analysis May 16, 2011 Tuesday, May 17, 2011 Outline • Review of the problem presented in the paper: “A comparison of angled sagittal MRI and conventional MRI in the diagnosis of herniated disc and stenosis in the cervical foramen” (Authors: Shim JH, Park CK, Lee JH, et. al) • Approach to solve this problem • Data Acquisition • Analysis Methods • Results • Discussion/Conclusions Tuesday, May 17, 2011 Review of Problem • Difficult to identify herniated discs and spinal stenosis using conventional (2D) MRI techniques • These conventional methods result in patients condition being misdiagnosed. “Conventional MRI”: Images acquired along one of three anatomical planes Tuesday, May 17, 2011 3D reconstructive CT Axial, T2-weighted Image: Image shows that the Cervical Foramen is cervical foramina are directed at 45 degrees directed downward around with respect to coronal 10-15 degrees with plane. respect to axial plane Tuesday, May 17, 2011 Orientation of Images Conventional MRI: Sagittal Protocol Oblique MRI: Sagittal Protocol Tuesday, May 17, 2011 Timeline • Week 1 (4/11-4/16) • Work on developing MR imaging protocols and sequences • Recruit volunteers (~4-5 volunteers) • Week 2 (4/17-4/23) • Continue developing imaging sequences and begin data acquisition at the MRI facility • Assisted by Dr. Deoni • Week 3&4 (4/24-5/7) • Continue developing and testing sequence • 4/27/2011: Acquisition of first subject • Mid Project Presentation: Describe the imaging protocols, present data that had been acquired from previous week, describe what still needs to be done. -

Unified Framework for Development, Deployment and Robust Testing Of
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Boise State University - ScholarWorks Boise State University ScholarWorks Computer Science Faculty Publications and Department of Computer Science Presentations 3-1-2011 Unified rF amework for Development, Deployment and Robust Testing of Neuroimaging Algorithms Alark Joshi Boise State University Dustin Scheinost Yale University Hirohito Okuda GE Healthcare Dominique Belhachemi Yale University Isabella Murphy Yale University See next page for additional authors This is an author-produced, peer-reviewed version of this article. The final publication is available at www.springerlink.com. Copyright restrictions may apply. DOI: 10.1007/s12021-010-9092-8 Authors Alark Joshi, Dustin Scheinost, Hirohito Okuda, Dominique Belhachemi, Isabella Murphy, Lawrence H. Staib, and Xenophon Papademetris This article is available at ScholarWorks: http://scholarworks.boisestate.edu/cs_facpubs/5 Unified framework for development, deployment and robust testing of neuroimaging algorithms Alark Joshi · Dustin Scheinost · Hirohito Okuda · Dominique Belhachemi · Isabella Murphy · Lawrence H. Staib · Xenophon Papademetris Received: date / Accepted: date Abstract Developing both graphical and command- 1 Introduction line user interfaces for neuroimaging algorithms requires considerable effort. Neuroimaging algorithms can meet Image analysis algorithms are typically developed to their potential only if they can be easily and frequently address a particular problem within a specific domain used by their intended users. Deployment of a large (functional MRI, cardiac, image-guided intervention plan- suite of such algorithms on multiple platforms requires ning and monitoring, etc.). Many of these algorithms consistency of user interface controls, consistent results are rapidly prototyped and developed without consid- across various platforms and thorough testing. -

MITK-Modelfit: a Generic Open-Source Framework for Model Fits and Their Exploration in Medical Imaging – Design, Implementatio
MITK-ModelFit: A generic open-source framework for model fits and their exploration in medical imaging – design, implementation and application on the example of DCE-MRI Charlotte Debus1-5,*,#, Ralf Floca5,6,*,#, Michael Ingrisch7, Ina Kompan5,6, Klaus Maier-Hein5,6,8, Amir Abdollahi1-5, and Marco Nolden6 1German Cancer Consortium (DKTK), Heidelberg, Germany 2Translational Radiation Oncology, German Cancer Research Center (DKFZ), Heidelberg, Germany 3Department of Radiation Oncology, Heidelberg Ion-Beam Therapy Center (HIT), Heidelberg University Hospital, Heidelberg, Germany 4National Center for Tumor Diseases (NCT), Heidelberg, Germany 5Heidelberg Institute of Radiation Oncology (HIRO), Germany 6Division of Medical Image Computing, German Cancer Research Center DKFZ, Germany 7Department of Radiology, University Hospital Munich, Ludwig-Maximilians-University Munich, Germany 8Section Pattern Recognition, Department of Radiation Oncology, Heidelberg University Hospital, Heidelberg, Germany # Correspondence: Charlotte Debus, PhD Department of Translational Radiation Oncology Heidelberg Ion-Beam Therapy Center (HIT) Im Neuenheimer Feld 450 69120 Heidelberg, Germany Email: [email protected] Phone: +49 6221 6538281 Ralf Floca, PhD Division of Medical Image Computing German Cancer Research Center (DKFZ) Im Neuenheimer Feld 280 69120 Heidelberg, Germany Email: [email protected] Phone: + 49 6221 42 2560 * Shared first-authors 1 Abstract Background: Many medical imaging techniques utilize fitting approaches for quantitative parameter estimation and analysis. Common examples are pharmacokinetic modeling in dynamic contrast- enhanced (DCE) magnetic resonance imaging (MRI)/computed tomography (CT), apparent diffusion coefficient calculations and intravoxel incoherent motion modeling in diffusion-weighted MRI and Z- spectra analysis in chemical exchange saturation transfer MRI. Most available software tools are limited to a special purpose and do not allow for own developments and extensions. -

Survey of Databases Used in Image Processing and Their Applications
International Journal of Scientific & Engineering Research Volume 2, Issue 10, Oct-2011 1 ISSN 2229-5518 Survey of Databases Used in Image Processing and Their Applications Shubhpreet Kaur, Gagandeep Jindal Abstract- This paper gives review of Medical image database (MIDB) systems which have been developed in the past few years for research for medical fraternity and students. In this paper, I have surveyed all available medical image databases relevant for research and their use. Keywords: Image database, Medical Image Database System. —————————— —————————— 1. INTRODUCTION Measurement and recording techniques, such as electroencephalography, magnetoencephalography Medical imaging is the technique and process used to (MEG), Electrocardiography (EKG) and others, can create images of the human for clinical purposes be seen as forms of medical imaging. Image Analysis (medical procedures seeking to reveal, diagnose or is done to ensure database consistency and reliable examine disease) or medical science. As a discipline, image processing. it is part of biological imaging and incorporates radiology, nuclear medicine, investigative Open source software for medical image analysis radiological sciences, endoscopy, (medical) Several open source software packages are available thermography, medical photography and for performing analysis of medical images: microscopy. ImageJ 3D Slicer ITK Shubhpreet Kaur is currently pursuing masters degree OsiriX program in Computer Science and engineering in GemIdent Chandigarh Engineering College, Mohali, India. E-mail: MicroDicom [email protected] FreeSurfer Gagandeep Jindal is currently assistant processor in 1.1 Images used in Medical Research department Computer Science and Engineering in Here is the description of various modalities that are Chandigarh Engineering College, Mohali, India. E-mail: used for the purpose of research by medical and [email protected] engineering students as well as doctors. -

An Open-Source Research Platform for Image-Guided Therapy
Int J CARS DOI 10.1007/s11548-015-1292-0 ORIGINAL ARTICLE CustusX: an open-source research platform for image-guided therapy Christian Askeland1,3 · Ole Vegard Solberg1 · Janne Beate Lervik Bakeng1 · Ingerid Reinertsen1 · Geir Arne Tangen1 · Erlend Fagertun Hofstad1 · Daniel Høyer Iversen1,2,3 · Cecilie Våpenstad1,2 · Tormod Selbekk1,3 · Thomas Langø1,3 · Toril A. Nagelhus Hernes2,3 · Håkon Olav Leira2,3 · Geirmund Unsgård2,3 · Frank Lindseth1,2,3 Received: 3 July 2015 / Accepted: 31 August 2015 © The Author(s) 2015. This article is published with open access at Springerlink.com Abstract Results The validation experiments show a navigation sys- Purpose CustusX is an image-guided therapy (IGT) research tem accuracy of <1.1mm, a frame rate of 20 fps, and latency platform dedicated to intraoperative navigation and ultra- of 285ms for a typical setup. The current platform is exten- sound imaging. In this paper, we present CustusX as a robust, sible, user-friendly and has a streamlined architecture and accurate, and extensible platform with full access to data and quality process. CustusX has successfully been used for algorithms and show examples of application in technologi- IGT research in neurosurgery, laparoscopic surgery, vascular cal and clinical IGT research. surgery, and bronchoscopy. Methods CustusX has been developed continuously for Conclusions CustusX is now a mature research platform more than 15years based on requirements from clinical for intraoperative navigation and ultrasound imaging and is and technological researchers within the framework of a ready for use by the IGT research community. CustusX is well-defined software quality process. The platform was open-source and freely available at http://www.custusx.org. -

Medical Image Processing Software
Wohlers Report 2018 Medical Image Processing Software Medical image Patient-specific medical devices and anatomical models are almost always produced using radiological imaging data. Medical image processing processing software is used to translate between radiology file formats and various software AM file formats. Theoretically, any volumetric radiological imaging dataset by Andy Christensen could be used to create these devices and models. However, without high- and Nicole Wake quality medical image data, the output from AM can be less than ideal. In this field, the old adage of “garbage in, garbage out” definitely applies. Due to the relative ease of image post-processing, computed tomography (CT) is the usual method for imaging bone structures and contrast- enhanced vasculature. In the dental field and for oral- and maxillofacial surgery, in-office cone-beam computed tomography (CBCT) has become popular. Another popular imaging technique that can be used to create anatomical models is magnetic resonance imaging (MRI). MRI is less useful for bone imaging, but its excellent soft tissue contrast makes it useful for soft tissue structures, solid organs, and cancerous lesions. Computed tomography: CT uses many X-ray projections through a subject to computationally reconstruct a cross-sectional image. As with traditional 2D X-ray imaging, a narrow X-ray beam is directed to pass through the subject and project onto an opposing detector. To create a cross-sectional image, the X-ray source and detector rotate around a stationary subject and acquire images at a number of angles. An image of the cross-section is then computed from these projections in a post-processing step. -
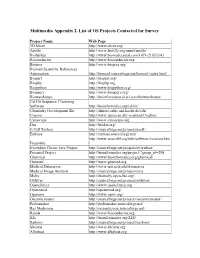
PDF File, 212 KB
Multimedia Appendix 2. List of OS Projects Contacted for Survey Project Name Web Page 3D Slicer http://www.slicer.org/ Apollo http://www.fruitfly.org/annot/apollo/ Biobuilder http://www.biomedcentral.com/1471-2105/5/43 Bioconductor http://www.bioconductor.org Biojava http://www.biojava.org/ Biomail Scientific References Automation http://biomail.sourceforge.net/biomail/index.html Bioperl http://bioperl.org/ Biophp http://biophp.org Biopython http://www.biopython.org/ Bioquery http://www.bioquery.org/ Biowarehouse http://bioinformatics.ai.sri.com/biowarehouse/ Cd-Hit Sequence Clustering Software http://bioinformatics.org/cd-hit/ Chemistry Development Kit http://almost.cubic.uni-koeln.de/cdk/ Coasim http://www.daimi.au.dk/~mailund/CoaSim/ Cytoscape http://www.cytoscape.org Das http://biodas.org/ E-Cell System http://sourceforge.net/projects/ecell/ Emboss http://emboss.sourceforge.net/ http://www.ensembl.org/info/software/versions.htm Ensemble l Eviewbox Dicom Java Project http://sourceforge.net/projects/eviewbox/ Freemed Project http://bioinformatics.org/project/?group_id=298 Ghemical http://www.bioinformatics.org/ghemical/ Gnumed http://www.gnumed.org Medical Dataserver http://www.mii.ucla.edu/dataserver Medical Image Analysis http://sourceforge.net/projects/mia Moby http://biomoby.open-bio.org/ Olduvai http://sourceforge.net/projects/olduvai/ Openclinica http://www.openclinica.org Openemed http://openemed.org/ Openemr http://www.oemr.org/ Oscarmcmaster http://sourceforge.net/projects/oscarmcmaster/ Probemaker http://probemaker.sourceforge.net/ -

Evaluation of DICOM Viewer Software for Workflow Integration in Clinical Trials
Evaluation of DICOM Viewer Software for Workflow Integration in Clinical Trials Daniel Haak1*, Charles-E. Page, Klaus Kabino, Thomas M. Deserno Department of Medical Informatics, Uniklinik RWTH Aachen, 52057 Aachen, Germany ABSTRACT The digital imaging and communications in medicine (DICOM) protocol is nowadays the leading standard for capture, exchange and storage of image data in medical applications. A broad range of commercial, free, and open source software tools supporting a variety of DICOM functionality exists. However, different from patient’s care in hospital, DICOM has not yet arrived in electronic data capture systems (EDCS) for clinical trials. Due to missing integration, even just the visualization of patient’s image data in electronic case report forms (eCRFs) is impossible. Four increasing levels for integration of DICOM components into EDCS are conceivable, raising functionality but also demands on interfaces with each level. Hence, in this paper, a comprehensive evaluation of 27 DICOM viewer software projects is performed, investigating viewing functionality as well as interfaces for integration. Concerning general, integration, and viewing requirements the survey involves the criteria (i) license, (ii) support, (iii) platform, (iv) interfaces, (v) two- dimensional (2D) and (vi) three-dimensional (3D) image viewing functionality. Optimal viewers are suggested for applications in clinical trials for 3D imaging, hospital communication, and workflow. Focusing on open source solutions, the viewers ImageJ and MicroView are superior for 3D visualization, whereas GingkoCADx is advantageous for hospital integration. Concerning workflow optimization in multi-centered clinical trials, we suggest the open source viewer Weasis. Covering most use cases, an EDCS and PACS interconnection with Weasis is suggested. -

Kitware Source Issue 21
SOFTWARE DEVELOPER’S QUARTERLY Issue 21 • April 2012 Editor’s Note ........................................................................... 1 PARAVIEW 3.14 RELEASED In late February, Kitware and the ParaView team released Recent Releases ..................................................................... 1 ParaView 3.14. This release features usability enhancements, improvements to the Plugin framework, new panels, and ParaView in Immersive Environments .................................. 3 more than 100 other resolved issues. Teaching Open Source .......................................................... 4 ParaView 3.14 features a redesigned Find Data dialog and Color Editor. The updated Find Data dialog makes it possible ParaView Query Selection Framework................................. 7 to use complex queries to select elements, including combin- ing multiple test cases with Boolean operations. The Color Ginkgo CADx: Open Source DICOM CADx Environment .... 8 Editor, used to edit lookup tables or color tables for scalar mapping, now enables independent editing of the color and Code Review, Topic Branches and VTK ................................. 9 opacity functions. Video Analysis on Business Intelligence, ParaView’s charting capabilities have been extended with a Studies in Computational Intelligence ............................... 11 new scatter plot matrix view and the ability to visualize mul- tiple dimensions of data in one compact form. This improved The Visible Patient .............................................................. -

Sveučilište U Zagrebu, Fakultet Strojarstva I Brodogradnje
Primjena neuronskih mreža za segmentaciju tkiva u biomedicinskim snimkama pacijenata Delovski, Boris Master's thesis / Diplomski rad 2019 Degree Grantor / Ustanova koja je dodijelila akademski / stručni stupanj: University of Zagreb, Faculty of Mechanical Engineering and Naval Architecture / Sveučilište u Zagrebu, Fakultet strojarstva i brodogradnje Permanent link / Trajna poveznica: https://urn.nsk.hr/urn:nbn:hr:235:481058 Rights / Prava: In copyright Download date / Datum preuzimanja: 2021-10-07 Repository / Repozitorij: Repository of Faculty of Mechanical Engineering and Naval Architecture University of Zagreb SVEUČILIŠTE U ZAGREBU FAKULTET STROJARSTVA I BRODOGRADNJE DIPLOMSKI RAD Boris Delovski Zagreb, godina 2019 SVEUČILIŠTE U ZAGREBU FAKULTET STROJARSTVA I BRODOGRADNJE DIPLOMSKI RAD Primjena neuronskih mreža za segmentaciju tkiva u biomedicinskim snimkama pacijenata Mentori: Student: Prof. dr. sc. Bojan Jerbić, dipl. ing. Boris Delovski Dr. sc. Filip Šuligoj, dipl. ing. Zagreb, godina 2019. Izjavljujem da sam ovaj rad izradio samostalno koristeći znanja stečena tijekom studija i navedenu literaturu. Zahvaljujem se svom mentoru, profesoru Bojanu Jerbiću te asistentu Filipu Šuligoju koji su mi svojim stručnim savjetima pomogli oblikovati i realizirati ovaj rad. Zahvalio bih se na svom znanju koje sam stekao za vrijeme izrade ovog rada, te na pruţenoj prilici da svoju ideju sprovedem od početka do kraja. TakoĎer bih se ţelio zahvaliti svojoj obitelji i prijateljima na pruţenoj podršci i razumijevanju. Boris Delovski Boris Delovski Diplomski