Updates to Clostridium Difficile Spore Germination
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Amerithrax Investigative Summary
The United States Department of Justice AMERITHRAX INVESTIGATIVE SUMMARY Released Pursuant to the Freedom of Information Act Friday, February 19, 2010 TABLE OF CONTENTS I. THE ANTHRAX LETTER ATTACKS . .1 II. EXECUTIVE SUMMARY . 4 A. Overview of the Amerithrax Investigation . .4 B. The Elimination of Dr. Steven J. Hatfill as a Suspect . .6 C. Summary of the Investigation of Dr. Bruce E. Ivins . 6 D. Summary of Evidence from the Investigation Implicating Dr. Ivins . .8 III. THE AMERITHRAX INVESTIGATION . 11 A. Introduction . .11 B. The Investigation Prior to the Scientific Conclusions in 2007 . 12 1. Early investigation of the letters and envelopes . .12 2. Preliminary scientific testing of the Bacillus anthracis spore powder . .13 3. Early scientific findings and conclusions . .14 4. Continuing investigative efforts . 16 5. Assessing individual suspects . .17 6. Dr. Steven J. Hatfill . .19 7. Simultaneous investigative initiatives . .21 C. The Genetic Analysis . .23 IV. THE EVIDENCE AGAINST DR. BRUCE E. IVINS . 25 A. Introduction . .25 B. Background of Dr. Ivins . .25 C. Opportunity, Access and Ability . 26 1. The creation of RMR-1029 – Dr. Ivins’s flask . .26 2. RMR-1029 is the source of the murder weapon . 28 3. Dr. Ivins’s suspicious lab hours just before each mailing . .29 4. Others with access to RMR-1029 have been ruled out . .33 5. Dr. Ivins’s considerable skill and familiarity with the necessary equipment . 36 D. Motive . .38 1. Dr. Ivins’s life’s work appeared destined for failure, absent an unexpected event . .39 2. Dr. Ivins was being subjected to increasing public criticism for his work . -

Investigation of Anthrax Associated with Intentional Exposure
October 19, 2001 / Vol. 50 / No. 41 889 Update: Investigation of Anthrax Associated with Intentional Exposure and Interim Public Health Guidelines, October 2001 893 Recognition of Illness Associated with the Intentional Release of a Biologic Agent 897 Weekly Update: West Nile Virus Activity — United States, October 10–16, 2001 Update: Investigation of Anthrax Associated with Intentional Exposure and Interim Public Health Guidelines, October 2001 On October 4, 2001, CDC and state and local public health authorities reported a case of inhalational anthrax in Florida (1 ). Additional cases of anthrax subsequently have been reported from Florida and New York City. This report updates the findings of these case investigations, which indicate that infections were caused by the intentional release of Bacillus anthracis. This report also includes interim guidelines for postexposure pro- phylaxis for prevention of inhalational anthrax and other information to assist epidemi- ologists, clinicians, and laboratorians responding to intentional anthrax exposures. For these investigations, a confirmed case of anthrax was defined as 1) a clinically compatible case of cutaneous, inhalational, or gastrointestinal illness* that is laboratory confirmed by isolation of B. anthracis from an affected tissue or site or 2) other labora- tory evidence of B. anthracis infection based on at least two supportive laboratory tests. A suspected case was defined as 1) a clinically compatible case of illness without isola- tion of B. anthracis and no alternative diagnosis, but with laboratory evidence of B. anthracis by one supportive laboratory test or 2) a clinically compatible case of an- thrax epidemiologically linked to a confirmed environmental exposure, but without cor- roborative laboratory evidence of B. -

2001 Anthrax Letters
Chapter Five: 2001 Anthrax Letters Author’s Note: The analysis and comments regarding the communication efforts described in this case study are solely those of the authors; this analysis does not represent the official position of the FDA. This case was selected because it is one of the few major federal efforts to distribute medical countermeasures in response to an acute biological incident. These events occurred more than a decade ago and represent the early stages of US biosecurity preparedness and response; however, this incident serves as an excellent illustration of the types of communication challenges expected in these scenarios. Due in part to the extended time since these events and the limited accessibility of individual communications and messages, this case study does not provide a comprehensive assessment of all communication efforts. In contrast to the previous case studies in this casebook, the FDA’s role in the 2001 anthrax response was relatively small, and as such, this analysis focuses principally on the communication efforts of the CDC and state and local public health agencies. The 2001 anthrax attacks have been studied extensively, and the myriad of internal and external assessments led to numerous changes to response and communications policies and protocols. The authors intend to use this case study as a means of highlighting communication challenges strictly within the context of this incident, not to evaluate the success or merit of changes made as a result of these events. Abstract The dissemination of Bacillus anthracis via the US Postal Service (USPS) in 2001 represented a new public health threat, the first intentional exposure to anthrax in the United States. -

Bacillus Cereus Pneumonia
‘Advances in Medicine and Biology’, Volume 67, 2013 Nova Science Publisher Inc., Editor: Berhardt LV To Alessia, my queen, and Giorgia, my princess Chapter BACILLUS CEREUS PNEUMONIA Vincenzo Savini* Clinical Microbiology and Virology, Spirito Santo Hospital, Pescara (PE), Italy ABSTRACT Bacillus cereus is a Gram positive/Gram variable environmental rod, that is emerging as a respiratory pathogen; particularly, pneumonia it causes may be serious and can resemble the anthrax disease. In fact, the organism is strictly related to the famous Bacillus anthracis, with which it shares genotypical, fenotypical, and pathogenic features. Treatment of B. cereus lower airway infections is becoming increasingly hard, due to the spread of multidrug resistance traits among members of the species. Hence, the present chapter’s scope is to shed a light on this bacterium’s lung pathogenicity, by depicting salient microbiological, epidemiological and clinical features it shows. Also, we would like to explore virulence determinants and resistance mechanisms that make B. cereus a life-threatening, potentially difficult-to-treat agent of airway pathologies. * Email: [email protected]. 2 Vincenzo Savini INTRODUCTION The genus Bacillus includes spore-forming species, strains of which are not usually considered to be clinically relevant when isolated from human specimens; in fact, these bacteria are known to be ubiquitously distributed in the environment and may easily contaminate culture material along with improperly handled sample collection devices (Miller, 2012; Brooks, 2001). Bacillus anthracis is the prominent agent of human pathologies within the genus Bacillus, although it is uncommon in most clinical laboratories (Miller, 2012). It is a frank pathogen, as it causes skin and enteric infections; above all, however, it is responsible for a serious lung disease (the ‘anthrax’) that is frequently associated with bloodstream infection and a high mortality rate (Frankard, 2004). -

Lessons from the Anthrax Attacks Implications for US
Lessons from the Anthrax Attacks Implications for US. Bioterrorism Preparedness A Report on a National Forum on Biodefense Author David Heymart Research Assistants Srusha Ac h terb erg, L Joelle Laszld Organized by the Center for Strategic and International Studies and the Defense Threat Reduction Agency --CFr”V? --.....a DlSTRf BUTION This report ISfor official use only; distribution authorized to U S. government agencies, designated contractors, and those with an official need Contains information that may be exempt from public release under the Freedom of Information Act. exemption number 2 (5 USC 552); exemption number 3 (’lo USC 130).Approvalof the Defense Threat Reduction Agency prior to public release is requrred Contract Number OTRAM-02-C-0013 For Official Use Only About CSIS For four decades, the Center for Stravgic and Internahonal Studies (CSIS)has been dedicated 10 providing world leaders with strategic insights on-and pohcy solubons tcurrentand emergtng global lssues CSIS IS led by John J Hamre, former L S deputy secretary of defense It is guided by a board of trustees chaired by former U S senator Sam Nunn and consistlng of prominent individuals horn both the public and private sectors The CSIS staff of 190 researchers and support staff focus pnrnardy on three subjecr areas First, CSIS addresses the fuU spectrum of new challenges to national and mternabonal security Second, it maintains resident experts on all of the world's myor geographical regions Third, it IS committed to helping to develop new methods of governance for the global age, to this end, CSIS has programs on technology and pubhc policy, International trade and finance, and energy Headquartered in Washington, D-C ,CSIS IS pnvate, bipartlsan, and tax-exempt CSIS docs not take specific policy positions, accordygly, all views expressed herein should be understood to be solely those ofthe author Spousor. -
Anthrax Reporting and Investigation Guideline
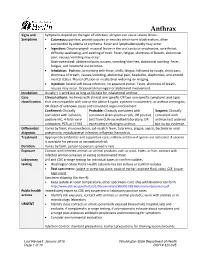
Anthrax Signs and Symptoms depend on the type of infection; all types can cause severe illness: Symptoms • Cutaneous: painless, pruritic papules or vesicles which form black eschars, often surrounded by edema or erythema. Fever and lymphadenopathy may occur. • Ingestion: Oropharyngeal: mucosal lesion in the oral cavity or oropharynx, sore throat, difficulty swallowing, and swelling of neck. Fever, fatigue, shortness of breath, abdominal pain, nausea/vomiting may occur. Gastrointestinal: abdominal pain, nausea, vomiting/diarrhea, abdominal swelling. Fever, fatigue, and headache are common. • Inhalation: Biphasic, presenting with fever, chills, fatigue, followed by cough, chest pain, shortness of breath, nausea/vomiting, abdominal pain, headache, diaphoresis, and altered mental status. Pleural effusion or mediastinal widening on imaging. • Injection: Severe soft tissue infection; no apparent eschar. Fever, shortness of breath, nausea may occur. Occasional meningeal or abdominal involvement. Incubation Usually < 1 week but as long as 60 days for inhalational anthrax Case Clinical criteria: An illness with at least one specific OR two non-specific symptoms and signs classification that are compatible with one of the above 4 types, systemic involvement, or anthrax meningitis; OR death of unknown cause and consistent organ involvement Confirmed: Clinically Probable: Clinically consistent with Suspect: Clinically consistent with isolation, consistent Gram-positive rods, OR positive consistent with positive IHC, 4-fold rise in test from CLIA-accredited laboratory, OR anthrax test ordered antibodies, PCR, or LF MS epi evidence relating to anthrax but no epi evidence Differential Varies by form; mononucleosis, cat-scratch fever, tularemia, plague, sepsis, bacterial or viral diagnosis pneumonia, mycobacterial infection, influenza, hantavirus Treatment Appropriate antibiotics and supportive care; anthrax antitoxin if spores are activated. -

Bioterrorism, Biological Weapons and Anthrax
Bioterrorism, Biological Weapons and Anthrax Part IV Written by Arthur H. Garrison Criminal Justice Planning Coordinator Delaware Criminal Justice Council Bioterrorism and biological weapons The use of bio-terrorism and bio-warfare dates back to 6th century when the Assyrians poisoned the well water of their enemies. The goal of using biological weapons is to cause massive sickness or death in the intended target. Bioterrorism and biological weapons The U.S. took the threat of biological weapons attack seriously after Gulf War. Anthrax vaccinations of U.S. troops Investigating Iraq and its biological weapons capacity The Soviet Union manufactured various types of biological weapons during the 1980’s • To be used after a nuclear exchange • Manufacturing new biological weapons – Gene engineering – creating new types of viruses/bacteria • Contagious viruses – Ebola, Marburg (Filoviruses) - Hemorrhagic fever diseases (vascular system dissolves) – Smallpox The spread of biological weapons after the fall of the Soviet Union •Material • Knowledge and expertise •Equipment Bioterrorism and biological weapons There are two basic categories of biological warfare agents. Microorganisms • living organic germs, such as anthrax (bacillus anthrax). –Bacteria –Viruses Toxins • By-products of living organisms (natural poisons) such as botulism (botulinum toxin) which is a by- product of growing the microorganism clostridium botulinum Bioterrorism and biological weapons The U.S. was a leader in the early research on biological weapons Research on making -

Antibacterial Photosensitization Through Activation of PNAS PLUS Coproporphyrinogen Oxidase
Antibacterial photosensitization through activation of PNAS PLUS coproporphyrinogen oxidase Matthew C. Surdela, Dennis J. Horvath Jr.a, Lisa J. Lojeka, Audra R. Fullena, Jocelyn Simpsona, Brendan F. Dutterb,c, Kenneth J. Sallenga, Jeremy B. Fordd, J. Logan Jenkinsd, Raju Nagarajane, Pedro L. Teixeiraf, Matthew Albertollec,g, Ivelin S. Georgieva,e,h, E. Duco Jansend, Gary A. Sulikowskib,c, D. Borden Lacya,d, Harry A. Daileyi,j,k, and Eric P. Skaara,1 aDepartment of Pathology, Microbiology, and Immunology, Vanderbilt University Medical Center, Nashville, TN 37232; bDepartment of Chemistry, Vanderbilt University, Nashville, TN 37232; cVanderbilt Institute for Chemical Biology, Nashville, TN 37232; dDepartment of Biomedical Engineering, Vanderbilt University, Nashville, TN 37232; eVanderbilt Vaccine Center, Vanderbilt University Medical Center, Nashville, TN 37232; fBiomedical Informatics, Vanderbilt University School of Medicine, Nashville, TN 37203; gDepartment of Biochemistry, Vanderbilt University, Nashville, TN 37232; hDepartment of Electrical Engineering and Computer Science, Vanderbilt University, Nashville, TN 37232; iBiomedical and Health Sciences Institute, University of Georgia, Athens, GA 30602; jDepartment of Microbiology, University of Georgia, Athens, GA 30602; and kDepartment of Biochemistry and Molecular Biology, University of Georgia, Athens, GA 30602 Edited by Ferric C. Fang, University of Washington School of Medicine, Seattle, WA, and accepted by Editorial Board Member Carl F. Nathan June 26, 2017 (received for review January 10, 2017) Gram-positive bacteria cause the majority of skin and soft tissue Small-molecule VU0038882 (‘882) was previously identified in infections (SSTIs), resulting in the most common reason for clinic a screen for activators of the S. aureus heme-sensing system two- visits in the United States. -

Characterization of an Endolysin Targeting Clostridioides Difficile
International Journal of Molecular Sciences Article Characterization of an Endolysin Targeting Clostridioides difficile That Affects Spore Outgrowth Shakhinur Islam Mondal 1,2 , Arzuba Akter 3, Lorraine A. Draper 1,4 , R. Paul Ross 1 and Colin Hill 1,4,* 1 APC Microbiome Ireland, University College Cork, T12 YT20 Cork, Ireland; [email protected] (S.I.M.); [email protected] (L.A.D.); [email protected] (R.P.R.) 2 Genetic Engineering and Biotechnology Department, Shahjalal University of Science and Technology, Sylhet 3114, Bangladesh 3 Biochemistry and Molecular Biology Department, Shahjalal University of Science and Technology, Sylhet 3114, Bangladesh; [email protected] 4 School of Microbiology, University College Cork, T12 K8AF Cork, Ireland * Correspondence: [email protected] Abstract: Clostridioides difficile is a spore-forming enteric pathogen causing life-threatening diarrhoea and colitis. Microbial disruption caused by antibiotics has been linked with susceptibility to, and transmission and relapse of, C. difficile infection. Therefore, there is an urgent need for novel therapeutics that are effective in preventing C. difficile growth, spore germination, and outgrowth. In recent years bacteriophage-derived endolysins and their derivatives show promise as a novel class of antibacterial agents. In this study, we recombinantly expressed and characterized a cell wall hydrolase (CWH) lysin from C. difficile phage, phiMMP01. The full-length CWH displayed lytic activity against selected C. difficile strains. However, removing the N-terminal cell wall binding domain, creating CWH351—656, resulted in increased and/or an expanded lytic spectrum of activity. C. difficile specificity was retained versus commensal clostridia and other bacterial species. -

Gao-15-80, Anthrax
United States Government Accountability Office Report to Congressional Requesters December 2014 ANTHRAX Agency Approaches to Validation and Statistical Analyses Could Be Improved GAO-15-80 December 2014 ANTHRAX Agency Approaches to Validation and Statistical Analyses Could Be Improved Highlights of GAO-15-80, a report to congressional requesters Why GAO Did This Study What GAO Found In 2001, the FBI investigated an After the 2001 Anthrax attacks, the genetic tests that were conducted by the intentional release of B. anthracis, a Federal Bureau of Investigation’s (FBI) four contractors were generally bacterium that causes anthrax, which scientifically verified and validated, and met the FBI’s criteria. However, GAO was identified as the Ames strain. found that the FBI lacked a comprehensive approach—or framework—that could Subsequently, FBI contractors have ensured standardization of the testing process. As a result, each of the developed and validated several contractors developed their tests differently, and one contractor did not conduct genetic tests to analyze B. anthracis verification testing, a key step in determining whether a test will meet a user’s samples for the presence of certain requirements, such as for sensitivity or accuracy. Also, GAO found that the genetic mutations. The FBI had contractors did not develop the level of statistical confidence for interpreting the previously collected and maintained testing results for the validation tests they performed. Responses to future these samples in a repository. incidents could be improved by using a standardized framework for achieving GAO was asked to review the FBI’s minimum performance standards during verification and validation, and by genetic test development process and incorporating statistical analyses when interpreting validation testing results. -

Anti-Anthrax Agents Means That New Antibiotics Are Needed to the 14-Membered Ring
research highlights NATURAL PRODUCTS infection and its resistance to treatment system — along with a dichloro group on Anti-anthrax agents means that new antibiotics are needed to the 14-membered ring. Fenical and the Angew. Chem. Int. Ed. 52, 7822–7824 (2013) target this bacterium. team decided to incorporate this dichloro In an effort to discover new antimicrobial moiety into anthracimycin to see what H H scaffolds, a team led by William Fenical at the effect this would have on the antimicrobial University of California at San Diego, USA, activity. Interestingly, the dichloro analogue H H H isolated metabolites from a marine organism was approximately half as potent against H H Cl H (a species of Streptomyces) and tested them Bacillus anthracis, however, it exhibited O O O O Cl for antibacterial activity. One metabolite greater activity against some strains of O O H O O showed potent activity against the Gram- Gram-negative bacteria — which the team positive Bacillus anthracis and methicillin- believe is linked to easier penetration Anthracimycin Dichloro analogue resistant Staphylococcus aureus; although through the cell wall. RJ it had only very limited activity against Few diseases carry the sinister connotations Gram-negative bacteria such as Escherichia REACTIVE INTERMEDIATES of anthrax. It is a deadly infection that coli. A series of NMR spectroscopy, mass Non-classical crystals predominantly affects herbivores, although spectrometry and X-ray crystallography Science 341, 62–64 (2013) it has also been used for bioterrorism analyses enabled the team to characterize and adapted for biowarfare. Anthrax is the structure and stereochemistry of this When either enantiomer of exo-2-norbornyl caused by Bacillus anthracis, and, like other new antimicrobial compound — which they bromide reacts with a nucleophile, a racemic bacterial diseases, it is normally treated with termed anthracimycin. -

Equine Anthrax
Equine Anthrax JUNE 2015 Cause Bacillus anthracis bacteria Risk of Exposure in Low Illinois Risk of High Transmission to exposed people Mode of Ingestion or inhalation of spores; handling contaminated carcass or Transmission hair Incubation Human: Cutaneous form: 3-10 days Period Inhalation form: 1-5 days Gastrointestinal form: 2-5 days Animal: 3-7 days (can range from 1-20 days) Cutaneous form accounts for most human cases-red, raised Clinical Signs- lesion; blister Human Pulmonary form- fever; vague sense of ill-being; muscle pain; cough; respiratory distress, sweating, shock, death Gastrointestinal form- fever, vomiting, bloody diarrhea; general ill-being Clinical Signs- Common symptom septicemia with enteritis and colic; bloody Animal diarrhea; edematous lesions especially on throat and neck; subcutaneous swellings; animals may die within 1-3 days, but can survive up to one week *Failure to achieve rigor mortis after death Control and Prevention Vaccinate livestock in endemic areas; Vaccinate individuals in high risk occupations; deep burial/burn infected carcass Comments If anthrax is suspected, do NOT perform a necropsy; reportable disease in Illinois; potential bioterrorist agent Additional Information http://emergency.cdc.gov/agent/anthrax/index.asp http://www.cfsph.iastate.edu/Factsheets/pdfs/anthrax.pdf http://www.cfsph.iastate.edu/FastFacts/pdfs/anthrax_F.pdf Equine Brucellosis JUNE 2015 Cause Brucella spp. bacteria Risk of Exposure in Low (Illinois is currently Brucellosis free) Illinois Risk of Transmission High to exposed people Mode of Contact with infected animals especially aborted fetuses, Transmission uterine fluids or membranes, and urine; inhalation or ingestion; contact with objects capable of harboring bacteria Incubation Human: 1 week- several months after infection Period Animal: Variable Fever; headache; chills; generalized weakness; nausea; Clinical Signs- weight loss; enlarged lymph nodes and spleen.