Efficient SNP Based Heritability Estimation and Multiple Phenotype
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Package 'Woodburymatrix'
Package ‘WoodburyMatrix’ August 10, 2020 Title Fast Matrix Operations via the Woodbury Matrix Identity Version 0.0.1 Description A hierarchy of classes and methods for manipulating matrices formed implic- itly from the sums of the inverses of other matrices, a situation commonly encountered in spa- tial statistics and related fields. Enables easy use of the Woodbury matrix identity and the ma- trix determinant lemma to allow computation (e.g., solving linear systems) without hav- ing to form the actual matrix. More information on the underlying linear alge- bra can be found in Harville, D. A. (1997) <doi:10.1007/b98818>. URL https://github.com/mbertolacci/WoodburyMatrix BugReports https://github.com/mbertolacci/WoodburyMatrix/issues License MIT + file LICENSE Encoding UTF-8 LazyData true RoxygenNote 7.1.1 Imports Matrix, methods Suggests covr, lintr, testthat, knitr, rmarkdown VignetteBuilder knitr NeedsCompilation no Author Michael Bertolacci [aut, cre, cph] (<https://orcid.org/0000-0003-0317-5941>) Maintainer Michael Bertolacci <[email protected]> Repository CRAN Date/Publication 2020-08-10 12:30:02 UTC R topics documented: determinant,WoodburyMatrix,logical-method . .2 instantiate . .2 mahalanobis . .3 normal-distribution-methods . .4 1 2 instantiate solve-methods . .5 WoodburyMatrix . .6 WoodburyMatrix-class . .8 Index 10 determinant,WoodburyMatrix,logical-method Calculate the determinant of a WoodburyMatrix object Description Calculates the (log) determinant of a WoodburyMatrix using the matrix determinant lemma. Usage ## S4 method for signature 'WoodburyMatrix,logical' determinant(x, logarithm) Arguments x A object that is a subclass of WoodburyMatrix logarithm Logical indicating whether to return the logarithm of the matrix. Value Same as base::determinant. -

Fast and Accurate Camera Covariance Computation for Large 3D Reconstruction
Fast and Accurate Camera Covariance Computation for Large 3D Reconstruction Michal Polic1, Wolfgang F¨orstner2, and Tomas Pajdla1 [0000−0003−3993−337X][0000−0003−1049−8140][0000−0001−6325−0072] 1 CIIRC, Czech Technical University in Prague, Czech Republic fmichal.polic,[email protected], www.ciirc.cvut.cz 2 University of Bonn, Germany [email protected], www.ipb.uni-bonn.de Abstract. Estimating uncertainty of camera parameters computed in Structure from Motion (SfM) is an important tool for evaluating the quality of the reconstruction and guiding the reconstruction process. Yet, the quality of the estimated parameters of large reconstructions has been rarely evaluated due to the computational challenges. We present a new algorithm which employs the sparsity of the uncertainty propagation and speeds the computation up about ten times w.r.t. previous approaches. Our computation is accurate and does not use any approximations. We can compute uncertainties of thousands of cameras in tens of seconds on a standard PC. We also demonstrate that our approach can be ef- fectively used for reconstructions of any size by applying it to smaller sub-reconstructions. Keywords: uncertainty, covariance propagation, Structure from Mo- tion, 3D reconstruction 1 Introduction Three-dimensional reconstruction has a wide range of applications (e.g. virtual reality, robot navigation or self-driving cars), and therefore is an output of many algorithms such as Structure from Motion (SfM), Simultaneous location and mapping (SLAM) or Multi-view Stereo (MVS). Recent work in SfM and SLAM has demonstrated that the geometry of three-dimensional scene can be obtained from a large number of images [1],[14],[16]. -

No Belief Propagation Required
Article The International Journal of Robotics Research No belief propagation required: Belief 1–43 © The Author(s) 2017 Reprints and permissions: space planning in high-dimensional state sagepub.co.uk/journalsPermissions.nav DOI: 10.1177/0278364917721629 spaces via factor graphs, the matrix journals.sagepub.com/home/ijr determinant lemma, and re-use of calculation Dmitry Kopitkov1 and Vadim Indelman2 Abstract We develop a computationally efficient approach for evaluating the information-theoretic term within belief space plan- ning (BSP), where during belief propagation the state vector can be constant or augmented. We consider both unfocused and focused problem settings, whereas uncertainty reduction of the entire system or only of chosen variables is of interest, respectively. State-of-the-art approaches typically propagate the belief state, for each candidate action, through calcula- tion of the posterior information (or covariance) matrix and subsequently compute its determinant (required for entropy). In contrast, our approach reduces runtime complexity by avoiding these calculations. We formulate the problem in terms of factor graphs and show that belief propagation is not needed, requiring instead a one-time calculation that depends on (the increasing with time) state dimensionality, and per-candidate calculations that are independent of the latter. To that end, we develop an augmented version of the matrix determinant lemma, and show that computations can be re-used when eval- uating impact of different candidate actions. These two key ingredients and the factor graph representation of the problem result in a computationally efficient (augmented) BSP approach that accounts for different sources of uncertainty and can be used with various sensing modalities. -

Ensemble Kalman Filter with Perturbed Observations in Weather Forecasting and Data Assimilation
Ensemble Kalman Filter with perturbed observations in weather forecasting and data assimilation Yihua Yang 28017913 Department of Mathematics and Statistics University of Reading April 10, 2020 Abstract Data assimilation in Mathematics provides algorithms for widespread applications in various fields, especially in meteorology. It is of practical use to deal with a large amount of information in several dimensions in the complex system that is hard to estimate. Weather forecasting is one of the applications, where the prediction of meteorological data are corrected given the observations. Numerous approaches are contained in data assimilation. One specific sequential method is the Kalman Filter. The core is to estimate unknown information with the new data that is measured and the prior data that is predicted. The main benefit of the Kalman Filter lies in the more accurate prediction than that with single measurements. As a matter of fact, there are different improved methods in the Kalman Filter. In this project, the Ensemble Kalman Filter with perturbed observations is considered. It is achieved by Monte Carlo simulation. In this method, the ensemble is involved in the calculation instead of the state vectors. In addition, the measurement with perturbation is viewed as the suitable observation. These changes compared with the Linear Kalman Filter make it more advantageous in that applications are not restricted in linear systems any more and less time is taken when the data are calculated by computers. The more flexibility as well as the less pressure of calculation enable the specific method to be frequently utilised than other methods. This project seeks to develop the Ensemble Kalman Filter with perturbed obser- vations gradually. -

Doctoral Thesis Probabilistic Data Reconciliation in Material Flow
Doctoral Thesis Probabilistic Data Reconciliation in Material Flow Analysis submitted in satisfaction of the requirements for the degree of Doctor of Science in Civil Engineering of the TU Wien, Faculty of Civil Engineering Dissertation Probabilistischer Datenausgleich in der Materialflussanalyse ausgeführt zum Zwecke der Erlangung des akademischen Grades eines Doktors der technischen Wissenschaft eingereicht an der Technischen Universität Wien, Fakultät für Bauingenieurwesen von Dipl.-Ing. Oliver Cencic Matr.Nr.: 09140006 Petronell-C., Austria Supervisor: Univ.-Prof. Dipl.-Ing. Dr.techn. Helmut Rechberger Institute for Water Quality and Resource Management Technische Universität Wien, Austria Supervisor: Univ.-Doz. Dipl.-Ing. Dr. Rudolf Frühwirth Institute of High Energy Physics (HEPHY) Austrian Academy of Sciences, Austria Assessor: Associate Prof. Reinout Heijungs Department of Econometrics and Operations Research Vrije Universiteit Amsterdam, The Netherlands Assessor: Prof. Julian M. Allwood Department of Engineering University of Cambridge, United Kingdom Vienna, February 2018 Acknowledgement First of all, I would like to express my deepest gratitude to Dr. Rudolf Frühwirth for his patience and his unlimited support during the work on this theses. I learned so much about statistics and programming in MATLAB through all the discussion we had. It was motivating, inspiring, and a great honor to work with him. I also want to thank Prof. Helmut Rechberger for supervising my work and giving me the freedom to work on mathematical methodologies deviating from the actual field of research at the department. I would also like to express my appreciation to Prof. Reinout Heijungs and Prof. Julian Allwood for their evaluation of my thesis. Special thanks to my colleagues Nada Dzubur, Oliver Schwab, David Laner and Manuel Hahn for giving me the chance to internally discuss uncertainty matters after many years of solitary work on this topic. -

The Sherman Morrison Iteration
The Sherman Morrison Iteration Joseph T. Slagel Thesis submitted to the Faculty of the Virginia Polytechnic Institute and State University in partial fulfillment of the requirements for the degree of Master of Science in Mathematics Julianne Chung, Chair Matthias Chung, Chair Serkan Gugercin May 6, 2015 Blacksburg, Virginia Keywords: \Sherman Morrison, regularized least squares problem, extremely underdetermined linear system," Copyright 2015, Joseph T. Slagel The Sherman Morrison Iteration Joseph T. Slagel (ABSTRACT) The Sherman Morrison iteration method is developed to solve regularized least squares problems. Notions of pivoting and splitting are deliberated on to make the method more robust. The Sherman Morrison iteration method is shown to be effective when dealing with an extremely underdetermined least squares problem. The performance of the Sherman Morrison iteration is compared to classic direct methods, as well as iterative methods, in a number of experiments. Specific Matlab implementation of the Sherman Morrison iteration is discussed, with Matlab codes for the method available in the appendix. Acknowledgments I acknowledge Dr. Julianne Chung and Dr. Matthias Chung for their masterful patience and sage-like guidance during the composition of this thesis. I also acknowledge Dr. Serkan Gugercin for serving on my committee and being a transcendent teacher. iii Contents 1 Introduction 1 2 Preliminaries 2 2.1 Ill-Posed Inverse Problems . .2 2.2 Least Squares Problems . .3 2.3 Tikhonov Regularization . .7 3 The Sherman Morrison Iteration 10 3.1 Developing the General Case . 10 3.1.1 Pivoting . 14 3.1.2 Splitting . 16 3.2 Application to Regularized Least Squares Problem . 22 3.3 Efficiencies . -
Arxiv:1407.4552V3 [Math.CO] 17 Feb 2016 Tk = Vr (1) R(K − 1) = Λ(V − 1) (2) Tk(K − 1) = Λv(V − 1) (3)
An Asymptotic Formula for the Number of Balanced Incomplete Block Design Incidence Matrices Aaron M. Montgomery Department of Mathematics and Computer Science, Baldwin Wallace University July 24, 2021 Abstract We identify a relationship between a random walk on a certain Euclidean lattice and incidence matrices of balanced incomplete block designs, which arise in combinatorial design theory. We then compute the return probability of the random walk and use it to obtain the asymptotic number of BIBD incidence matrices (as the number of columns increases). Our strategy is similar in spirit to the one used by de Launey and Levin to count partial Hadamard matrices. 1 Introduction In this paper, we will explore a relationship between a particular family of random walks and a certain collection of combinatorially-defined matrices. These matrices, known as balanced incomplete block design incidence matrices, are important in combinatorial design theory. Our goal will be to estimate the number of these matrices as the number of columns increases. Although this task is quite difficult to do directly, it is possible to estimate the return probability of the random walk and then to exploit the connection between the two problems to estimate the number of the matrices. This tactic, adopted from [6], is noteworthy because it does not require explicit construction of the matrices being studied. Definition 1. We say that a v × t matrix populated with 1's and 0's is an incidence matrix of a balanced incomplete block design if there are positive integers k and λ such that: • each column has exactly k 1's, and • each pair of distinct rows has inner product λ, which is independent of the choice of the pair. -

Tensor Algebra, Linear Algebra, Matrix Algebra, Multilinear Algebra
Appendix A Tensor Algebra, Linear Algebra, Matrix Algebra, Multilinear Algebra The key word “algebra” is derived from the name and the work of the nineth-century scientist Mohammed ibn Mˆusˆa al-Khowˆarizmi who was born in what is now Uzbekistan and worked in Bagdad at the court of Harun al-Rashid’s son.“al-jabr” appears in the title of his book Kitab al-jabr wail muqabala where he discusses symbolic methods for the solution of equations, K. Vogel: Mohammed Ibn Musa Alchwarizmi’s Algorismus; Das forested Lehrbuch zum Rechnen mit indischen Ziffern, 1461, Otto Zeller Verlagsbuchhandlung, Aalen 1963). Accordingly what is an algebra and how is it tied to the notion of a vector space,atensor space, respectively? By an algebra we mean asetS of elements and a finite set M of operations. Each operation .opera/k is a single-valued function assigning to every finite ordered sequence .x1;:::;xn/ of n D n.k/ elements of S a value S .opera/k .x1;:::;xk / D xl in . In particular for .opera/k.x1;x2/ the operation is called binary,for.opera/k.x1;x2;x3/ ternary, in general for .opera/k.x1;:::;xn/n-ary. For a given set of operation symbols .opera/1, .opera/2;:::, .opera/k we define a word.Inlinear algebra the set M has basically two elements, namely two internal relations .opera/1 worded “addition” (including inverse addition: subtraction) and .opera/2 worded “multiplication” (including inverse multiplication: division). Here the elements of the set S are vectors over the field R of real numbers as long as we refer to linear algebra.Incontrast,in multilinear algebra the elements of the set S are tensors over the field of real numbers R. -

Towards More Efficient SPSD Matrix Approximation and CUR Matrix Decomposition
Journal of Machine Learning Research 17 (2016) 1-49 Submitted 5/15; Revised 4/16; Published 12/16 Towards More Efficient SPSD Matrix Approximation and CUR Matrix Decomposition Shusen Wang [email protected] Department of Statistics University of California at Berkeley Berkeley, CA 94720, USA Zhihua Zhang [email protected] School of Mathematical Sciences Peking University Beijing 100871, China Tong Zhang [email protected] Department of Statistics Rutgers University Piscataway, New Jersey 08854, USA Editor: Gert Lanckriet Abstract Symmetric positive semi-definite (SPSD) matrix approximation methods have been extensively used to speed up large-scale eigenvalue computation and kernel learning methods. The standard sketch based method, which we call the prototype model, produces relatively accurate approximations, but is inefficient on large square matrices. The Nystr¨om method is highly efficient, but can only achieve low accuracy. In this paper we propose a novel model that we call the fast SPSD matrix approximation model. The fast model is nearly as efficient as the Nystr¨ommethod and as accurate as the prototype model. We show that the fast model can potentially solve eigenvalue problems and kernel learning problems in linear time with respect to the matrix size n to achieve 1 + relative-error, whereas both the prototype model and the Nystr¨ommethod cost at least quadratic time to attain comparable error bound. Empirical comparisons among the prototype model, the Nystr¨ommethod, and our fast model demonstrate the superiority of the fast model. We also contribute new understandings of the Nystr¨ommethod. The Nystr¨ommethod is a special instance of our fast model and is approximation to the prototype model. -

Invertible Matrix - Wikipedia, the Free Encyclopedia
Invertible matrix - Wikipedia, the free encyclopedia http://en.wikipedia.org/wiki/Invertible_matrix Invertible matrix From Wikipedia, the free encyclopedia In linear algebra an n-by-n (square) matrix A is called invertible (some authors use nonsingular or nondegenerate) if there exists an n-by-n matrix B such that where In denotes the n-by-n identity matrix and the multiplication used is ordinary matrix multiplication. If this is the case, then the matrix B is uniquely determined by A and is called the inverse of A, denoted by A−1. It follows from the theory of matrices that if for finite square matrices A and B, then also [1] Non-square matrices (m-by-n matrices for which m ≠ n) do not have an inverse. However, in some cases such a matrix may have a left inverse or right inverse. If A is m-by-n and the rank of A is equal to n, then A has a left inverse: an n-by-m matrix B such that BA = I. If A has rank m, then it has a right inverse: an n-by-m matrix B such that AB = I. A square matrix that is not invertible is called singular or degenerate. A square matrix is singular if and only if its determinant is 0. Singular matrices are rare in the sense that if you pick a random square matrix over a continuous uniform distribution on its entries, it will almost surely not be singular. While the most common case is that of matrices over the real or complex numbers, all these definitions can be given for matrices over any commutative ring. -
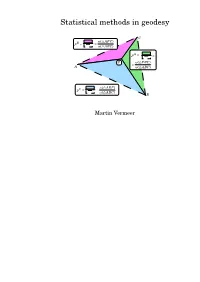
Statistical Methods in Geodesy
Statistical methods in geodesy C !( APC) pB 4 Æ Æ !( ABC) 4 A p = Æ P !( PBC) 4 A Æ !( ABC) 4 !( ABP) pC 4 Æ Æ !( ABC) 4 B Martin Vermeer Contents Contents iii 1. Free network and datum1 1.1 Theory............................. 1 1.2 Example: a levelling network ............... 2 1.3 Fixing the datum....................... 4 1.4 Other examples........................ 9 2. Similarity transformations (S-transformations) and cri- terion matrices 13 2.1 Complex co-ordinates and point variances . 13 2.2 S-transformation in the complex plane.......... 14 2.3 Standard form of the criterion matrix .......... 14 2.4 S-transformation of the criterion matrix......... 17 2.5 S-transformations as members of a group . 18 2.6 The S-transformation of variances ............ 21 3. The affine S-transformation 23 3.1 Triangulation and the finite elements method . 23 3.2 Bilinear affine transformation............... 23 3.3 Applying the method of affine transformation in a lo- cal situation.......................... 26 3.4 A theoretical analysis of error propagation . 26 3.5 The case of height measurement.............. 29 4. Determining the shape of an object (circle, sphere, straight line) 31 – iii – iv Contents 4.1 The general case....................... 31 4.2 Example: circle........................ 32 4.3 Exercises............................ 34 5. 3D network, industrial measurements with a system of several theodolites 37 5.1 Three dimensional theodolite measurement (EPLA) . 37 5.2 Example............................ 38 5.3 Different ways to create the scale............. 40 6. Deformation analysis 41 6.1 One dimensional deformation analysis.......... 41 6.2 Two dimensional deformation analysis.......... 42 6.3 Example........................... -

On Computing Sparse Generalized Inverses and Sparse-Inverse/Low-Rank Decompositions by Victor K. Fuentes a Dissertation Submitte
On Computing Sparse Generalized Inverses and Sparse-Inverse/Low-Rank Decompositions by Victor K. Fuentes A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Industrial and Operations Engineering) in the University of Michigan 2019 Doctoral Committee: Professor Jon Lee, Chair Assistant Professor Laura Balzano Professor Marina Epelman Professor Marcia Fampa Victor K. Fuentes [email protected] ORCID iD: 0000-0002-9874-554X © Victor K. Fuentes 2019 Dedication I dedicate my dissertation to Betty S. Keesey and Maureen Keesey Fuentes. ii Acknowledgments There are many people that have earned my gratitude and appreciation for their contributions to my time in graduate school. More specifically, I would like to extend thanks to a number of groups of people, without whom this thesis (and graduate experience) would not have been in the slightest bit possible. First, I am indebted to my advisor, Jon Lee, for providing an experience unlike any else that has changed me for the better. On an academic level, Jon provided an environment with which to explore what it means to be researcher, one which allowed me the opportunity to both develop my un- derstanding of the vastness that is the field of optimization and experience firsthand the difficulties that come with the territory. On a personal level,I found (and continue to find) myself fortunate to have a person such as Jon to serve as inspiration during my time here, providing a sounding board on how to configure (and then re-configure) the delicate balance of beinga graduate student, a researcher, and your own person (as well as how those are not necessarily mutually exclusive).