Exploration of Microbial Diversity and Evolution Through Cultivation Independent Phylogenomics
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Chemical Structures of Some Examples of Earlier Characterized Antibiotic and Anticancer Specialized
Supplementary figure S1: Chemical structures of some examples of earlier characterized antibiotic and anticancer specialized metabolites: (A) salinilactam, (B) lactocillin, (C) streptochlorin, (D) abyssomicin C and (E) salinosporamide K. Figure S2. Heat map representing hierarchical classification of the SMGCs detected in all the metagenomes in the dataset. Table S1: The sampling locations of each of the sites in the dataset. Sample Sample Bio-project Site depth accession accession Samples Latitude Longitude Site description (m) number in SRA number in SRA AT0050m01B1-4C1 SRS598124 PRJNA193416 Atlantis II water column 50, 200, Water column AT0200m01C1-4D1 SRS598125 21°36'19.0" 38°12'09.0 700 and above the brine N "E (ATII 50, ATII 200, 1500 pool water layers AT0700m01C1-3D1 SRS598128 ATII 700, ATII 1500) AT1500m01B1-3C1 SRS598129 ATBRUCL SRS1029632 PRJNA193416 Atlantis II brine 21°36'19.0" 38°12'09.0 1996– Brine pool water ATBRLCL1-3 SRS1029579 (ATII UCL, ATII INF, N "E 2025 layers ATII LCL) ATBRINP SRS481323 PRJNA219363 ATIID-1a SRS1120041 PRJNA299097 ATIID-1b SRS1120130 ATIID-2 SRS1120133 2168 + Sea sediments Atlantis II - sediments 21°36'19.0" 38°12'09.0 ~3.5 core underlying ATII ATIID-3 SRS1120134 (ATII SDM) N "E length brine pool ATIID-4 SRS1120135 ATIID-5 SRS1120142 ATIID-6 SRS1120143 Discovery Deep brine DDBRINP SRS481325 PRJNA219363 21°17'11.0" 38°17'14.0 2026– Brine pool water N "E 2042 layers (DD INF, DD BR) DDBRINE DD-1 SRS1120158 PRJNA299097 DD-2 SRS1120203 DD-3 SRS1120205 Discovery Deep 2180 + Sea sediments sediments 21°17'11.0" -

The 2014 Golden Gate National Parks Bioblitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 ON THIS PAGE Photograph of BioBlitz participants conducting data entry into iNaturalist. Photograph courtesy of the National Park Service. ON THE COVER Photograph of BioBlitz participants collecting aquatic species data in the Presidio of San Francisco. Photograph courtesy of National Park Service. The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 Elizabeth Edson1, Michelle O’Herron1, Alison Forrestel2, Daniel George3 1Golden Gate Parks Conservancy Building 201 Fort Mason San Francisco, CA 94129 2National Park Service. Golden Gate National Recreation Area Fort Cronkhite, Bldg. 1061 Sausalito, CA 94965 3National Park Service. San Francisco Bay Area Network Inventory & Monitoring Program Manager Fort Cronkhite, Bldg. 1063 Sausalito, CA 94965 March 2016 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate comprehensive information and analysis about natural resources and related topics concerning lands managed by the National Park Service. -

Pinpointing the Origin of Mitochondria Zhang Wang Hanchuan, Hubei
Pinpointing the origin of mitochondria Zhang Wang Hanchuan, Hubei, China B.S., Wuhan University, 2009 A Dissertation presented to the Graduate Faculty of the University of Virginia in Candidacy for the Degree of Doctor of Philosophy Department of Biology University of Virginia August, 2014 ii Abstract The explosive growth of genomic data presents both opportunities and challenges for the study of evolutionary biology, ecology and diversity. Genome-scale phylogenetic analysis (known as phylogenomics) has demonstrated its power in resolving the evolutionary tree of life and deciphering various fascinating questions regarding the origin and evolution of earth’s contemporary organisms. One of the most fundamental events in the earth’s history of life regards the origin of mitochondria. Overwhelming evidence supports the endosymbiotic theory that mitochondria originated once from a free-living α-proteobacterium that was engulfed by its host probably 2 billion years ago. However, its exact position in the tree of life remains highly debated. In particular, systematic errors including sparse taxonomic sampling, high evolutionary rate and sequence composition bias have long plagued the mitochondrial phylogenetics. This dissertation employs an integrated phylogenomic approach toward pinpointing the origin of mitochondria. By strategically sequencing 18 phylogenetically novel α-proteobacterial genomes, using a set of “well-behaved” phylogenetic markers with lower evolutionary rates and less composition bias, and applying more realistic phylogenetic models that better account for the systematic errors, the presented phylogenomic study for the first time placed the mitochondria unequivocally within the Rickettsiales order of α- proteobacteria, as a sister clade to the Rickettsiaceae and Anaplasmataceae families, all subtended by the Holosporaceae family. -

“Candidatus Deianiraea Vastatrix” with the Ciliate Paramecium Suggests
bioRxiv preprint doi: https://doi.org/10.1101/479196; this version posted November 27, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The extracellular association of the bacterium “Candidatus Deianiraea vastatrix” with the ciliate Paramecium suggests an alternative scenario for the evolution of Rickettsiales 5 Castelli M.1, Sabaneyeva E.2, Lanzoni O.3, Lebedeva N.4, Floriano A.M.5, Gaiarsa S.5,6, Benken K.7, Modeo L. 3, Bandi C.1, Potekhin A.8, Sassera D.5*, Petroni G.3* 1. Centro Romeo ed Enrica Invernizzi Ricerca Pediatrica, Dipartimento di Bioscienze, Università 10 degli studi di Milano, Milan, Italy 2. Department of Cytology and Histology, Faculty of Biology, Saint Petersburg State University, Saint-Petersburg, Russia 3. Dipartimento di Biologia, Università di Pisa, Pisa, Italy 4 Centre of Core Facilities “Culture Collections of Microorganisms”, Saint Petersburg State 15 University, Saint Petersburg, Russia 5. Dipartimento di Biologia e Biotecnologie, Università degli studi di Pavia, Pavia, Italy 6. UOC Microbiologia e Virologia, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy 7. Core Facility Center for Microscopy and Microanalysis, Saint Petersburg State University, Saint- Petersburg, Russia 20 8. Department of Microbiology, Faculty of Biology, Saint Petersburg State University, Saint- Petersburg, Russia * Corresponding authors, contacts: [email protected] ; [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/479196; this version posted November 27, 2018. -

Ehrlichiosis and Anaplasmosis Are Tick-Borne Diseases Caused by Obligate Anaplasmosis: Intracellular Bacteria in the Genera Ehrlichia and Anaplasma
Ehrlichiosis and Importance Ehrlichiosis and anaplasmosis are tick-borne diseases caused by obligate Anaplasmosis: intracellular bacteria in the genera Ehrlichia and Anaplasma. These organisms are widespread in nature; the reservoir hosts include numerous wild animals, as well as Zoonotic Species some domesticated species. For many years, Ehrlichia and Anaplasma species have been known to cause illness in pets and livestock. The consequences of exposure vary Canine Monocytic Ehrlichiosis, from asymptomatic infections to severe, potentially fatal illness. Some organisms Canine Hemorrhagic Fever, have also been recognized as human pathogens since the 1980s and 1990s. Tropical Canine Pancytopenia, Etiology Tracker Dog Disease, Ehrlichiosis and anaplasmosis are caused by members of the genera Ehrlichia Canine Tick Typhus, and Anaplasma, respectively. Both genera contain small, pleomorphic, Gram negative, Nairobi Bleeding Disorder, obligate intracellular organisms, and belong to the family Anaplasmataceae, order Canine Granulocytic Ehrlichiosis, Rickettsiales. They are classified as α-proteobacteria. A number of Ehrlichia and Canine Granulocytic Anaplasmosis, Anaplasma species affect animals. A limited number of these organisms have also Equine Granulocytic Ehrlichiosis, been identified in people. Equine Granulocytic Anaplasmosis, Recent changes in taxonomy can make the nomenclature of the Anaplasmataceae Tick-borne Fever, and their diseases somewhat confusing. At one time, ehrlichiosis was a group of Pasture Fever, diseases caused by organisms that mostly replicated in membrane-bound cytoplasmic Human Monocytic Ehrlichiosis, vacuoles of leukocytes, and belonged to the genus Ehrlichia, tribe Ehrlichieae and Human Granulocytic Anaplasmosis, family Rickettsiaceae. The names of the diseases were often based on the host Human Granulocytic Ehrlichiosis, species, together with type of leukocyte most often infected. -
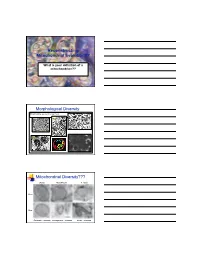
Reconstructing Mitochondrial Evolution?? Morphological Diversity Mitochondrial Diversity???
Reconstructing Mitochondrial Evolution?? What is your definition of a mitochondrion?? Morphological Diversity Mitochondria as we all know them: Suprarenal gland Liver cell Plasma cell Adrenal cortex Mitochondrial Diversity??? Chicken Neocallimastix T. foetus 100 nm 50 nm Entamoeba - mitosome microsporidian - mitosome Giardia - mitosome Reconstructing Evolution Mitochondrial evolution well established endosymbiotic theory α-proteobacterium - Rickettsia prowazekii Hydrogenosomal evolution No DNA NOW 2 examples Nyctotherus and Blastocystis (MLO) Several proteins similar to mitochondria Mitosome evolution No DNA Few proteins identified similar to mitochondria Origins via Endosymbiosis Current dogma - mitochondria and related Aerobic α-proteobacterium prokaryote gave rise to present day mitochondria. organelles arose just once in evolution Are hydrogenosomes and mitosomes of anaerobic protists derived from the same proto-mitochondrion? Evidence for: accumulating evidence for several proteins that are currently found in mitochondria - Proteins of Fe-S cluster formation. Scenario A Common ancestor facultative Scenario B organism? degenerate mitochondrion invoke lateral gene transfer from anaerobic prokaryotes Hypotheses for Mito Acquisition Mitochondrion-related Organelles 2 way to interpret this summary - can you think of both? Groups with mitochondrial homologues Hjort et al Phil. Trans. R. Soc. B 2010 365, 713-727 Origin of Hydrogenosomes (Mitosomes?) 1. a)Conversion of Mitochondria HYD b)Common ancestor with mitochondria ? HYD 2. Independent Origin α-proteobacterium ? - anaerobic bacterium HYD Organelles - origins and biogenesis Approaches: (1) Conduct phylogenetic analyses of similar proteins Hsp70 Fd Hsp60 Isc subunits (2) Examine protein targeting to the organelle matrix protein targeting membrane protein targeting (3) Characterize membrane/translocation components These components could have evolved as the endosymbiont was converted to organelle. Reveals evolutionary history. -

Table S4. Phylogenetic Distribution of Bacterial and Archaea Genomes in Groups A, B, C, D, and X
Table S4. Phylogenetic distribution of bacterial and archaea genomes in groups A, B, C, D, and X. Group A a: Total number of genomes in the taxon b: Number of group A genomes in the taxon c: Percentage of group A genomes in the taxon a b c cellular organisms 5007 2974 59.4 |__ Bacteria 4769 2935 61.5 | |__ Proteobacteria 1854 1570 84.7 | | |__ Gammaproteobacteria 711 631 88.7 | | | |__ Enterobacterales 112 97 86.6 | | | | |__ Enterobacteriaceae 41 32 78.0 | | | | | |__ unclassified Enterobacteriaceae 13 7 53.8 | | | | |__ Erwiniaceae 30 28 93.3 | | | | | |__ Erwinia 10 10 100.0 | | | | | |__ Buchnera 8 8 100.0 | | | | | | |__ Buchnera aphidicola 8 8 100.0 | | | | | |__ Pantoea 8 8 100.0 | | | | |__ Yersiniaceae 14 14 100.0 | | | | | |__ Serratia 8 8 100.0 | | | | |__ Morganellaceae 13 10 76.9 | | | | |__ Pectobacteriaceae 8 8 100.0 | | | |__ Alteromonadales 94 94 100.0 | | | | |__ Alteromonadaceae 34 34 100.0 | | | | | |__ Marinobacter 12 12 100.0 | | | | |__ Shewanellaceae 17 17 100.0 | | | | | |__ Shewanella 17 17 100.0 | | | | |__ Pseudoalteromonadaceae 16 16 100.0 | | | | | |__ Pseudoalteromonas 15 15 100.0 | | | | |__ Idiomarinaceae 9 9 100.0 | | | | | |__ Idiomarina 9 9 100.0 | | | | |__ Colwelliaceae 6 6 100.0 | | | |__ Pseudomonadales 81 81 100.0 | | | | |__ Moraxellaceae 41 41 100.0 | | | | | |__ Acinetobacter 25 25 100.0 | | | | | |__ Psychrobacter 8 8 100.0 | | | | | |__ Moraxella 6 6 100.0 | | | | |__ Pseudomonadaceae 40 40 100.0 | | | | | |__ Pseudomonas 38 38 100.0 | | | |__ Oceanospirillales 73 72 98.6 | | | | |__ Oceanospirillaceae -

Ticks and Tick-Borne Diseases 10 (2019) 1070–1077
Ticks and Tick-borne Diseases 10 (2019) 1070–1077 Contents lists available at ScienceDirect Ticks and Tick-borne Diseases journal homepage: www.elsevier.com/locate/ttbdis Original article Tissue tropism and metabolic pathways of Midichloria mitochondrii suggest tissue-specific functions in the symbiosis with Ixodes ricinus T Emanuela Olivieria,1, Sara Episb,c,1, Michele Castellib,c, Ilaria Varotto Boccazzib,c, ⁎ Claudia Romeod, Alessandro Desiròe, Chiara Bazzocchic,d,f, Claudio Bandib,c, Davide Sasseraa, a Department of Biology and Biotechnology, University of Pavia, via Ferrata 9, 27100, Pavia, Italy b Department of Biosciences University of Milan, Milan, Italy c Pediatric Clinical Research Center "Romeo ed Enrica Invernizzi”, University of Milan, 20133, Milan, Italy d Department of Veterinary Medicine, Università degli Studi di Milano, via Celoria 10, 20133, Milano, Italy e Department of Plant Soil and Microbial Sciences, Michigan State University, East Lansing, MI, USA f Coordinated Research Center "EpiSoMI", University of Milan, 20133, Milan, Italy ARTICLE INFO ABSTRACT Keywords: A wide range of arthropod species harbour bacterial endosymbionts in various tissues, many of them playing Midichloria mitochondrii important roles in the fitness and biology of their hosts. In several cases, many different symbionts have been Tick endosymbionts reported to coexist simultaneously within the same host and synergistic or antagonistic interactions can occur Nutrient provisioning between them. While the associations with endosymbiotic bacteria have been widely studied in many insect Energetic provisioning species, in ticks such interactions are less investigated. Anti-oxidative defence The females and immatures of Ixodes ricinus (Ixodidae), the most common hard tick in Europe, harbour the Osmotic regulation intracellular endosymbiont “Candidatus Midichloria mitochondrii” with a prevalence up to 100%, suggesting a mutualistic relationship. -

Table S5. the Information of the Bacteria Annotated in the Soil Community at Species Level
Table S5. The information of the bacteria annotated in the soil community at species level No. Phylum Class Order Family Genus Species The number of contigs Abundance(%) 1 Firmicutes Bacilli Bacillales Bacillaceae Bacillus Bacillus cereus 1749 5.145782459 2 Bacteroidetes Cytophagia Cytophagales Hymenobacteraceae Hymenobacter Hymenobacter sedentarius 1538 4.52499338 3 Gemmatimonadetes Gemmatimonadetes Gemmatimonadales Gemmatimonadaceae Gemmatirosa Gemmatirosa kalamazoonesis 1020 3.000970902 4 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas indica 797 2.344876284 5 Firmicutes Bacilli Lactobacillales Streptococcaceae Lactococcus Lactococcus piscium 542 1.594633558 6 Actinobacteria Thermoleophilia Solirubrobacterales Conexibacteraceae Conexibacter Conexibacter woesei 471 1.385742446 7 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas taxi 430 1.265115184 8 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas wittichii 388 1.141545794 9 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas sp. FARSPH 298 0.876754244 10 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sorangium cellulosum 260 0.764953367 11 Proteobacteria Deltaproteobacteria Myxococcales Polyangiaceae Sorangium Sphingomonas sp. Cra20 260 0.764953367 12 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas panacis 252 0.741416341 -

Dark Biological Superoxide Production As a Significant Flux and Sink of Marine Dissolved Oxygen
Dark biological superoxide production as a significant flux and sink of marine dissolved oxygen Kevin M. Sutherlanda,b, Scott D. Wankela, and Colleen M. Hansela,1 aDepartment of Marine Chemistry and Geochemistry, Woods Hole Oceanographic Institution, Woods Hole, MA 02543; and bDepartment of Earth, Atmospheric and Planetary Science, Massachusetts Institute of Technology, Cambridge, MA 02139 Edited by Donald E. Canfield, Institute of Biology and Nordic Center for Earth Evolution, University of Southern Denmark, Odense M., Denmark, and approved January 3, 2020 (received for review July 19, 2019) The balance between sources and sinks of molecular oxygen in the concentrations, even when the subject lacked a CO2 concentrating oceans has greatly impacted the composition of Earth’s atmo- mechanism (2). Studies of Mehler-related oxygen reduction in sphere since the evolution of oxygenic photosynthesis, thereby some cyanobacteria have been shown to exceed 40% of GOP (6, exerting key influence on Earth’s climate and the redox state of 7). Altogether, these studies demonstrate that Mehler-related (sub)surface Earth. The canonical source and sink terms of the oxygen loss in marine cyanobacteria and algae likely represents marine oxygen budget include photosynthesis, respiration, photo- a larger proportion of total nonrespiratory O2 reduction than is respiration, the Mehler reaction, and other smaller terms. How- observed in higher plants. ever, recent advances in understanding cryptic oxygen cycling, The role of intracellular superoxide production as a significant namely the ubiquitous one-electron reduction of O2 to superoxide sink of oxygen has been recognized since the 1950s for its place in by microorganisms outside the cell, remains unexplored as a po- the Mehler reaction (8); however, the role of extracellular su- tential player in global oxygen dynamics. -

The Tick Endosymbiont Candidatus Midichloria Mitochondrii And
Budachetri et al. Microbiome (2018) 6:141 https://doi.org/10.1186/s40168-018-0524-2 RESEARCH Open Access The tick endosymbiont Candidatus Midichloria mitochondrii and selenoproteins are essential for the growth of Rickettsia parkeri in the Gulf Coast tick vector Khemraj Budachetri1, Deepak Kumar1, Gary Crispell1, Christine Beck2, Gregory Dasch3 and Shahid Karim1* Abstract Background: Pathogen colonization inside tick tissues is a significant aspect of the overall competence of a vector. Amblyomma maculatum is a competent vector of the spotted fever group rickettsiae, Rickettsia parkeri. When R. parkeri colonizes its tick host, it has the opportunity to dynamically interact with not just its host but with the endosymbionts living within it, and this enables it to modulate the tick’s defenses by regulating tick gene expression. The microbiome in A. maculatum is dominated by two endosymbiont microbes: a Francisella-like endosymbiont (FLE) and Candidatus Midichloria mitochondrii (CMM). A range of selenium-containing proteins (selenoproteins) in A. maculatum ticks protects them from oxidative stress during blood feeding and pathogen infections. Here, we investigated rickettsial multiplication in the presence of tick endosymbionts and characterized the functional significance of selenoproteins during R. parkeri replication in the tick. Results: FLE and CMM were quantified throughout the tick life stages by quantitative PCR in R. parkeri-infected and uninfected ticks. R. parkeri infection was found to decrease the FLE numbers but CMM thrived across the tick life cycle. Our qRT-PCR analysis indicated that the transcripts of genes with functions related to redox (selenogenes) were upregulated in ticks infected with R. parkeri. Three differentially expressed proteins, selenoprotein M, selenoprotein O, and selenoprotein S were silenced to examine their functional significance during rickettsial replication within the tick tissues. -

Host-Adaptation in Legionellales Is 2.4 Ga, Coincident with Eukaryogenesis
bioRxiv preprint doi: https://doi.org/10.1101/852004; this version posted February 27, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 1 Host-adaptation in Legionellales is 2.4 Ga, 2 coincident with eukaryogenesis 3 4 5 Eric Hugoson1,2, Tea Ammunét1 †, and Lionel Guy1* 6 7 1 Department of Medical Biochemistry and Microbiology, Science for Life Laboratories, 8 Uppsala University, Box 582, 75123 Uppsala, Sweden 9 2 Department of Microbial Population Biology, Max Planck Institute for Evolutionary 10 Biology, D-24306 Plön, Germany 11 † current address: Medical Bioinformatics Centre, Turku Bioscience, University of Turku, 12 Tykistökatu 6A, 20520 Turku, Finland 13 * corresponding author 14 1 bioRxiv preprint doi: https://doi.org/10.1101/852004; this version posted February 27, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 15 Abstract 16 Bacteria adapting to living in a host cell caused the most salient events in the evolution of 17 eukaryotes, namely the seminal fusion with an archaeon 1, and the emergence of both the 18 mitochondrion and the chloroplast 2. A bacterial clade that may hold the key to understanding 19 these events is the deep-branching gammaproteobacterial order Legionellales – containing 20 among others Coxiella and Legionella – of which all known members grow inside eukaryotic 21 cells 3.