Toponym Resolution in Scientific Papers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Adaptive Geoparsing Method for Toponym Recognition and Resolution in Unstructured Text
remote sensing Article Adaptive Geoparsing Method for Toponym Recognition and Resolution in Unstructured Text Edwin Aldana-Bobadilla 1,∗ , Alejandro Molina-Villegas 2 , Ivan Lopez-Arevalo 3 , Shanel Reyes-Palacios 3, Victor Muñiz-Sanchez 4 and Jean Arreola-Trapala 4 1 Conacyt-Centro de Investigación y de Estudios Avanzados del I.P.N. (Cinvestav), Victoria 87130, Mexico; [email protected] 2 Conacyt-Centro de Investigación en Ciencias de Información Geoespacial (Centrogeo), Mérida 97302, Mexico; [email protected] 3 Centro de Investigación y de Estudios Avanzados del I.P.N. Unidad Tamaulipas (Cinvestav Tamaulipas), Victoria 87130, Mexico; [email protected] (I.L.); [email protected] (S.R.) 4 Centro de Investigación en Matemáticas (Cimat), Monterrey 66628, Mexico; [email protected] (V.M.); [email protected] (J.A.) * Correspondence: [email protected] Received: 10 July 2020; Accepted: 13 September 2020; Published: 17 September 2020 Abstract: The automatic extraction of geospatial information is an important aspect of data mining. Computer systems capable of discovering geographic information from natural language involve a complex process called geoparsing, which includes two important tasks: geographic entity recognition and toponym resolution. The first task could be approached through a machine learning approach, in which case a model is trained to recognize a sequence of characters (words) corresponding to geographic entities. The second task consists of assigning such entities to their most likely coordinates. Frequently, the latter process involves solving referential ambiguities. In this paper, we propose an extensible geoparsing approach including geographic entity recognition based on a neural network model and disambiguation based on what we have called dynamic context disambiguation. -

Georeferencing Accuracy of Geoeye-1 Stereo Imagery: Experiences in a Japanese Test Field
International Archives of the Photogrammetry, Remote Sensing and Spatial Information Science, Volume XXXVIII, Part 8, Kyoto Japan 2010 GEOREFERENCING ACCURACY OF GEOEYE-1 STEREO IMAGERY: EXPERIENCES IN A JAPANESE TEST FIELD Y. Meguro a, *, C.S.Fraser b a Japan Space Imaging Corp., 8-1Yaesu 2-Chome, Chuo-Ku, Tokyo 104-0028, Japan - [email protected] b Department of Geomatics, University of Melbourne VIC 3010, Australia - [email protected] Commission VIII, Working Group ICWG IV/VIII KEY WORDS: GeoEye-1, Stereo, RPCs, geopositioning, accuracy evaluation, bias compensation, satellite imagery ABSTRACT: High-resolution satellite imagery (HRSI) is being increasingly employed for large-scale topographic mapping, and especially for geodatabase updating. As the spatial resolution of HRSI sensors increases, so the potential georeferencing accuracy also improves. However, accuracy is not a function of spatial resolution alone, as it is also dependent upon radiometric image quality, the dynamics of the image scanning, and the fidelity of the sensor orientation model, both directly from orbit and attitude observations and indirectly from ground control points (GCPs). Users might anticipate accuracies of, say, 1 pixel in planimetry and 1-3 pixels in height when using GCPs. However, there are practical and in some cases administrative/legal imperatives for the georeferencing accuracy of HRSI systems to be quantified through well controlled tests. This paper discusses an investigation into the georeferencing accuracy attainable from the GeoEye-1 satellite, and specifically the 3D accuracy achievable from stereo imagery. Both direct georeferencing via supplied RPCs and indirect georeferencing via ground control and bias-corrected RPCs were examined for a stereo pair of pansharpened GeoEye-1 Basic images covering the Tsukuba Test Field in Japan, which contains more than 100 precisely surveyed and image identifiable GCPs. -

Knowledge-Driven Geospatial Location Resolution for Phylogeographic
Bioinformatics, 31, 2015, i348–i356 doi: 10.1093/bioinformatics/btv259 ISMB/ECCB 2015 Knowledge-driven geospatial location resolution for phylogeographic models of virus migration Davy Weissenbacher1,*, Tasnia Tahsin1, Rachel Beard1,2, Mari Figaro1,2, Robert Rivera1, Matthew Scotch1,2 and Graciela Gonzalez1 1Department of Biomedical Informatics, Arizona State University, Scottsdale, AZ 85259, USA and 2Center for Environmental Security, Biodesign Institute, Arizona State University, Tempe, AZ 85287-5904, USA *To whom correspondence should be addressed. Abstract Summary: Diseases caused by zoonotic viruses (viruses transmittable between humans and ani- mals) are a major threat to public health throughout the world. By studying virus migration and mutation patterns, the field of phylogeography provides a valuable tool for improving their surveil- lance. A key component in phylogeographic analysis of zoonotic viruses involves identifying the specific locations of relevant viral sequences. This is usually accomplished by querying public data- bases such as GenBank and examining the geospatial metadata in the record. When sufficient detail is not available, a logical next step is for the researcher to conduct a manual survey of the corresponding published articles. Motivation: In this article, we present a system for detection and disambiguation of locations (topo- nym resolution) in full-text articles to automate the retrieval of sufficient metadata. Our system has been tested on a manually annotated corpus of journal articles related to phylogeography using inte- grated heuristics for location disambiguation including a distance heuristic, a population heuristic and a novel heuristic utilizing knowledge obtained from GenBank metadata (i.e. a ‘metadata heuristic’). Results: For detecting and disambiguating locations, our system performed best using the meta- data heuristic (0.54 Precision, 0.89 Recall and 0.68 F-score). -

Introduction to Georeferencing
Introduction to Georeferencing Turning paper maps to interactive layers DMDS Workshop Jay Brodeur 2019-02-29 Today’s Outline ➢ Basic fundamentals of GIS and geospatial data ⚬ Vectors vs. rasters ⚬ Coordinate reference systems ➢ Introduction to Quantum GIS (QGIS) ➢ Hands-on Problem-Solving Assignments Quantum GIS (QGIS) ➢ Free and open-source GIS software ➢ User-friendly, fully-functional; relatively lightweight ➢ Product of the Open Source Geospatial Foundation (OSGeo) ➢ Built in C++; uses python for scripting and plugins ➢ Version 1.0 released in 2009 ➢ Current version: 3.16; Long-term release (LTR): 3.14 GDAL - Geospatial Data Abstraction Library www.gdal.org SAGA - System for Automated Geoscientific Analyses saga-gis.org/ GRASS - Geographic Resources Analysis Support System www.grass.osgeo.org QGIS - Quantum GIS qgis.org GeoTools www.geotools.org Helpful QGIS Tutorials and Resources ➢ QGIS Tutorials: http://www.qgistutorials.com/en/ ➢ QGIS Quicktips with Klas Karlsson: https://www.youtube.com/channel/UCxs7cfMwzgGZhtUuwhny4-Q ➢ QGIS Training Guide: https://docs.qgis.org/2.8/en/docs/training_manual/ Geospatial Data Fundamentals Representing real-world geographic information in a computer Task 1: Compare vector and raster data layers Objective: Download some openly-available raster and vector data. Explore the differences. Topics Covered: ➢ The QGIS Interface ➢ Geospatial data ➢ Layer styling ➢ Vectors vs rasters Online version of notes: https://goo.gl/H5vqNs Task 1.1: Downloading vector raster data & adding it to your map 1. Navigate a browser to Scholars Geoportal: http://geo.scholarsportal.info/ 2. Search for ‘index’ using the ‘Historical Maps’ category 3. Load the 1:25,000 topo map index 4. Use the interactive index to download the 1972 map sheet of Hamilton 5. -
Georeferencing
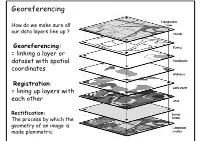
Georeferencing How do we make sure all our data layers line up ? Georeferencing: = linking a layer or dataset with spatial coordinates Registration: = lining up layers with each other Rectification: The process by which the geometry of an image is made planimetric Georeferencing • ‘To georeference’ the act of assigning locations to atoms of information • Is essential in GIS, since all information must be linked to the Earth’s surface • The method of georeferencing must be: – Unique, linking information to exactly one location – Shared, so different users understand the meaning of a georeference – Persistent through time, so today’s georeferences are still meaningful tomorrow Georeferencing Based on Data Types • Raster and Raster • Vector and Vector • Raster and Vector Geocoding Concepts and Definitions Definition of Geocoding • Geocoding can be broadly defined as the assignment of a code to a geographic location. Usually however, Geocoding refers to a more specific assignment of geographic coordinates (latitude,Longitude) to an individual address.. UN Report Definition of Geocoding • What is Geocoding • Geocoding is a process of creating map features from addresses, place names, or similar textual information based on attributes associated with a referenced geographic database, typically a street network that has address ranges associated with each street segment or 'link' running from one intersection to the next. Definition of Geocoding • What is Geocoding • Geocoding typically uses Interpolation as a method to find the location information about an address. – (If the address along one side of a block range from 1 to 199, then Street Number = 66 is about one-third of the way along that side of the block.) • Data required: – Reasonably clean, consistent list of legal addresses (i.e. -

A Comparison of Address Point and Street Geocoding Techniques
A COMPARISON OF ADDRESS POINT AND STREET GEOCODING TECHNIQUES IN A COMPUTER AIDED DISPATCH ENVIRONMENT by Jimmy Tuan Dao A Thesis Presented to the FACULTY OF THE USC GRADUATE SCHOOL UNIVERSITY OF SOUTHERN CALIFORNIA In Partial Fulfillment of the Requirements for the Degree MASTER OF SCIENCE (GEOGRAPHIC INFORMATION SCIENCE AND TECHNOLOGY) August 2015 Copyright 2015 Jimmy Tuan Dao DEDICATION I dedicate this document to my mother, sister, and Marie Knudsen who have inspired and motivated me throughout this process. The encouragement that I received from my mother and sister (Kim and Vanna) give me the motivation to pursue this master's degree, so I can better myself. I also owe much of this success to my sweetheart and life-partner, Marie who is always available to help review my papers and offer support when I was frustrated and wanting to quit. Thank you and I love you! ii ACKNOWLEDGMENTS I will be forever grateful to the faculty and classmates at the Spatial Science Institute for their support throughout my master’s program, which has been a wonderful period of my life. Thank you to my thesis committee members: Professors Darren Ruddell, Jennifer Swift, and Daniel Warshawsky for their assistance and guidance throughout this process. I also want to thank the City of Brea, my family, friends, and Marie Knudsen without whom I could not have made it this far. Thank you! iii TABLE OF CONTENTS DEDICATION ii ACKNOWLEDGMENTS iii LIST OF TABLES iii LIST OF FIGURES iv LIST OF ABBREVIATIONS vi ABSTRACT vii CHAPTER 1: INTRODUCTION 1 1.1 Brea, California -

TAGGS: Grouping Tweets to Improve Global Geotagging for Disaster Response Jens De Bruijn1, Hans De Moel1, Brenden Jongman1,2, Jurjen Wagemaker3, Jeroen C.J.H
Nat. Hazards Earth Syst. Sci. Discuss., https://doi.org/10.5194/nhess-2017-203 Manuscript under review for journal Nat. Hazards Earth Syst. Sci. Discussion started: 13 June 2017 c Author(s) 2017. CC BY 3.0 License. TAGGS: Grouping Tweets to Improve Global Geotagging for Disaster Response Jens de Bruijn1, Hans de Moel1, Brenden Jongman1,2, Jurjen Wagemaker3, Jeroen C.J.H. Aerts1 1Institute for Environmental Studies, VU University, Amsterdam, 1081HV, The Netherlands 5 2Global Facility for Disaster Reduction and Recovery, World Bank Group, Washington D.C., 20433, USA 3FloodTags, The Hague, 2511 BE, The Netherlands Correspondence to: Jens de Bruijn ([email protected]) Abstract. The availability of timely and accurate information about ongoing events is important for relief organizations seeking to effectively respond to disasters. Recently, social media platforms, and in particular Twitter, have gained traction as 10 a novel source of information on disaster events. Unfortunately, geographical information is rarely attached to tweets, which hinders the use of Twitter for geographical applications. As a solution, analyses of a tweet’s text, combined with an evaluation of its metadata, can help to increase the number of geo-located tweets. This paper describes a new algorithm (TAGGS), that georeferences tweets by using the spatial information of groups of tweets mentioning the same location. This technique results in a roughly twofold increase in the number of geo-located tweets as compared to existing methods. We applied this approach 15 to 35.1 million flood-related tweets in 12 languages, collected over 2.5 years. In the dataset, we found 11.6 million tweets mentioning one or more flood locations, which can be towns (6.9 million), provinces (3.3 million), or countries (2.2 million). -

Structured Toponym Resolution Using Combined Hierarchical Place Categories∗
In GIR’13: Proceedings of the 7th ACM SIGSPATIAL Workshop on Geographic Information Retrieval, Orlando, FL, November 2013 (GIR’13 Best Paper Award). Structured Toponym Resolution Using Combined Hierarchical Place Categories∗ Marco D. Adelfio Hanan Samet Center for Automation Research, Institute for Advanced Computer Studies Department of Computer Science, University of Maryland College Park, MD 20742 USA {marco, hjs}@cs.umd.edu ABSTRACT list or table is geotagged with the locations it references. To- ponym resolution and geotagging are common topics in cur- Determining geographic interpretations for place names, or rent research but the results can be inaccurate when place toponyms, involves resolving multiple types of ambiguity. names are not well-specified (that is, when place names are Place names commonly occur within lists and data tables, not followed by a geographic container, such as a country, whose authors frequently omit qualifications (such as city state, or province name). Our aim is to utilize the context of or state containers) for place names because they expect other places named within the table or list to disambiguate the meaning of individual place names to be obvious from place names that have multiple geographic interpretations. context. We present a novel technique for place name dis- ambiguation (also known as toponym resolution) that uses Geographic references are a very common component of Bayesian inference to assign categories to lists or tables data tables that can occur in both (1) the case where the containing place names, and then interprets individual to- primary entities of a table are geographic or (2) the case ponyms based on the most likely category assignments. -

A Survey on Geocoding: Algorithms and Datasets for Toponym Resolution
A Survey on Geocoding: Algorithms and Datasets for Toponym Resolution Anonymous ACL submission Abstract survey and critical evaluation of the currently avail- 040 able datasets, evaluation metrics, and geocoding 041 001 Geocoding, the task of converting unstructured algorithms. Our contributions are: 042 002 text to structured spatial data, has recently seen 003 progress thanks to a variety of new datasets, • the first survey to review deep learning ap- 043 004 evaluation metrics, and machine-learning algo- proaches to geocoding 044 005 rithms. We provide a comprehensive survey • comprehensive coverage of geocoding sys- 045 006 to review, organize and analyze recent work tems, which increased by 50% in the last 4 046 007 on geocoding (also known as toponym resolu- 008 tion) where the text is matched to geospatial years 047 009 coordinates and/or ontologies. We summarize • comprehensive coverage of annotated geocod- 048 010 the findings of this research and suggest some ing datasets, which increased by 100% in the 049 011 promising directions for future work. last 4 years 050 012 1 Introduction 2 Background 051 013 Geocoding, also called toponym resolution or to- An early work on geocoding, Amitay et al.(2004), 052 014 ponym disambiguation, is the subtask of geopars- identifies two important types of ambiguity: A 053 015 ing that disambiguates place names in text. The place name may also have a non-geographic mean- 054 016 goal of geocoding is, given a textual mention of a ing, such as Turkey the country vs. turkey the ani- 055 017 location, to choose the corresponding geospatial co- mal, and two places may have the same name, such 056 018 ordinates, geospatial polygon, or entry in a geospa- as the San Jose in California and the San Jose in 057 019 tial database. -

An Integrated INS/GPS Approach to the Georeferencing of Remotely
I PEER-REVIEWED ARTICLE An Integrated INSIGPS Approach to the Georeferencing of Remotely Sensed Data Abstract remote areas such as are found in many developing coun- A general model for the georeferencing of remotely sensed tries. data by an onboard positioning and orientation system is In the case of pushbroom imagery, the georeferencing presented as a problem of rigid body motion. The determina- problem is further complicated due to the need for the in- tion of the six independent parameters of motion by discrete stantaneous position and attitude information at each line of measurements from ineNealand satellife systems is directly imagery. Ground control alone is not sufficient to resolve all related to the problem of exterior orientation. The contribu- of the three position and three orientation (attitude) parame- tion of each measuring system to the determination of the ters associated with each exposure station, of which there three translational and three rotational parameters is treated may be thousands in a single mission. In order to overcome in detail, with emphasis on the contribution of inertial navi- these problems, systems have been proposed whereby multi- gation systems (INS] and single- and multi-antenna receivers ple look angles are simultaneously recorded with slight vari- of the Global Positioning System [GPS). The advantages of an ations in alignment (Hofmann et al., 1988). integrated INSIGPS approach are briefly discussed. Position- Another approach has been through the use of auxiliary ing and orientation accuracies obtainable from available position and navigation sensors which have been simulta- systems are then highlighted using selected results to em- neously flown during imaging missions. -

Guide to Best Practices for Georeferencing Includes Index ISBN: 87-92020-00-3
Guide to Best Practices for Georeferencing Guide to Best Practices for Georeferencing BioGeomancer Consortium August 2006 Published by: Global Biodiversity Information Facility, Copenhagen http://www.gbif.org Copyright © 2006 The Regents of the University of California. All rights reserved. The information in this book represents the professional opinion of the authors, and does not necessarily represent the views of the publisher or of the Regents of the University of California. While the authors and the publisher have attempted to make this book as accurate and as thorough as possible, the information contained herein is provided on an "As Is" basis, and without any warranties with respect to its accuracy or completeness. The authors, the publisher and the Regents of the University of California shall have no liability to any person or entity for any loss or damage caused by using the information provided in this book. Guide to Best Practices for Georeferencing Includes Index ISBN: 87-92020-00-3 Recommended Citation: Chapman, A.D. and J. Wieczorek (eds). 2006. Guide to Best Practices for Georeferencing. Copenhagen: Global Biodiversity Information Facility. Edited by: Arthur D. Chapman and John Wieczorek Contributors: J.Wieczorek, R.Guralnick, A.Chapman, C.Frazier, N.Rios, R.Beaman, Q.Guo. Contents CONTENTS ..............................................................................................................................................I GLOSSARY ...........................................................................................................................................III -

Using GIS with GPS
GIS Best Practices Using GIS with GPS March 2009 Table of Contents What Is GIS? 1 GPS-GAP Changes the Way We View the Earth 3 GIS and GPS Integration Eases Public Road Inventory 9 Dominion and Verizon Use Mobile GIS and GPS to Conduct Joint-Use Pole Survey 13 A Cost-Effective Approach to GPS/GIS Integration for Archaeological Surveying 17 Facilities Survey Feasible With GIS and GPS 23 i What Is GIS? Making decisions based on geography is basic to human thinking. Where shall we go, what will it be like, and what shall we do when we get there are applied to the simple event of going to the store or to the major event of launching a bathysphere into the ocean's depths. By understanding geography and people's relationship to location, we can make informed decisions about the way we live on our planet. A geographic information system (GIS) is a technological tool for comprehending geography and making intelligent decisions. GIS organizes geographic data so that a person reading a map can select data necessary for a specifi c project or task. A thematic map has a table of contents that allows the reader to add layers of information to a basemap of real-world locations. For example, a social analyst might use the basemap of Eugene, Oregon, and select datasets from the U.S. Census Bureau to add data layers to a map that shows residents' education levels, ages, and employment status. With an ability to combine a variety of datasets in an infi nite number of ways, GIS is a useful tool for nearly every fi eld of knowledge from archaeology to zoology.