Hereditary Hypotransferrinemia
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Orphanet Report Series Rare Diseases Collection
Marche des Maladies Rares – Alliance Maladies Rares Orphanet Report Series Rare Diseases collection DecemberOctober 2013 2009 List of rare diseases and synonyms Listed in alphabetical order www.orpha.net 20102206 Rare diseases listed in alphabetical order ORPHA ORPHA ORPHA Disease name Disease name Disease name Number Number Number 289157 1-alpha-hydroxylase deficiency 309127 3-hydroxyacyl-CoA dehydrogenase 228384 5q14.3 microdeletion syndrome deficiency 293948 1p21.3 microdeletion syndrome 314655 5q31.3 microdeletion syndrome 939 3-hydroxyisobutyric aciduria 1606 1p36 deletion syndrome 228415 5q35 microduplication syndrome 2616 3M syndrome 250989 1q21.1 microdeletion syndrome 96125 6p subtelomeric deletion syndrome 2616 3-M syndrome 250994 1q21.1 microduplication syndrome 251046 6p22 microdeletion syndrome 293843 3MC syndrome 250999 1q41q42 microdeletion syndrome 96125 6p25 microdeletion syndrome 6 3-methylcrotonylglycinuria 250999 1q41-q42 microdeletion syndrome 99135 6-phosphogluconate dehydrogenase 67046 3-methylglutaconic aciduria type 1 deficiency 238769 1q44 microdeletion syndrome 111 3-methylglutaconic aciduria type 2 13 6-pyruvoyl-tetrahydropterin synthase 976 2,8 dihydroxyadenine urolithiasis deficiency 67047 3-methylglutaconic aciduria type 3 869 2A syndrome 75857 6q terminal deletion 67048 3-methylglutaconic aciduria type 4 79154 2-aminoadipic 2-oxoadipic aciduria 171829 6q16 deletion syndrome 66634 3-methylglutaconic aciduria type 5 19 2-hydroxyglutaric acidemia 251056 6q25 microdeletion syndrome 352328 3-methylglutaconic -

Mackenzie's Mission Gene & Condition List
Mackenzie’s Mission Gene & Condition List What conditions are being screened for in Mackenzie’s Mission? Genetic carrier screening offered through this research study has been carefully developed. It is focused on providing people with information about their chance of having children with a severe genetic condition occurring in childhood. The screening is designed to provide genetic information that is relevant and useful, and to minimise uncertain and unclear information. How the conditions and genes are selected The Mackenzie’s Mission reproductive genetic carrier screen currently includes approximately 1300 genes which are associated with about 750 conditions. The reason there are fewer conditions than genes is that some genetic conditions can be caused by changes in more than one gene. The gene list is reviewed regularly. To select the conditions and genes to be screened, a committee comprised of experts in genetics and screening was established including: clinical geneticists, genetic scientists, a genetic pathologist, genetic counsellors, an ethicist and a parent of a child with a genetic condition. The following criteria were developed and are used to select the genes to be included: • Screening the gene is technically possible using currently available technology • The gene is known to cause a genetic condition • The condition affects people in childhood • The condition has a serious impact on a person’s quality of life and/or is life-limiting o For many of the conditions there is no treatment or the treatment is very burdensome for the child and their family. For some conditions very early diagnosis and treatment can make a difference for the child. -

Iron Deficiency Anemia (IRIDA) • Rare
Iron Deficiency: Review Melinda Wu, MD, MCR Oregon Health & Science University OHSU10/17/2019 Disclosure Information: a) Moderators/panelists/presenters: Melinda Wu has nothing to disclose. OHSUb) Funding sources: NIH/NHLBI- K08 HL133493 Objectives 1) To review iron body homeostasis 2) To review the etiologies of iron deficiency OHSU3) To review various treatment options of iron deficiency Part I: Review of Iron Body OHSUHomeostasis Iron Balance in the Body Iron is required for growth of all cells, not just hemoglobin! Heme proteins: cytochromes, catalase, peroxidase, cytochrome oxidase Flavoproteins: cytochrome C reductase, succinic dehydrogenase, NADH oxidase, xanthine oxidase Too little Too much Not enough for essential Accumulates in organs proteins: Promotes the formation of: • Hemoglobin • Oxygen radicals •OHSURibonucleotide reductase • Lipid peroxidation (DNA synthesis) • DNA damage • Cytochromes • Tissue fibrosis • Oxidases Iron Economy • The average adult has 4-5 g of body iron. • ~10% of dietary iron absorbed, exclusively in duodenum • Varies with: • Iron content of diet • Bioavailability of dietary iron • Iron stores in body • Erythropoietic demand • Hypoxia • Inflammation • More than half is incorporated into erythroid precursors/mature erythrocytes OHSU• Only ~1-2 mg of iron enters and leaves the body in a day on average. • About 1 mg of iron is lost daily in menstruating women. Lesjak, M.; K. S. Srai, S. Role of Dietary Flavonoids in Iron Homeostasis. Pharmaceuticals 2019 Systemic Iron Regulation: Absorption Iron status is regulated entirely at the level of absorption! • Heme iron (30-70%) > non-heme iron (<5%) • 2 stable oxidation states: Ferrous (Fe 2+) > Ferric (Fe 3+) • Elemental iron must be reduced to Fe2+ iron to be absorbed 1. -

Mechanisms of Copper Deficiency in the Zebrafish Embryo Erik Madsen Washington University in St
Washington University in St. Louis Washington University Open Scholarship All Theses and Dissertations (ETDs) January 2010 Mechanisms of Copper Deficiency in the Zebrafish Embryo Erik Madsen Washington University in St. Louis Follow this and additional works at: https://openscholarship.wustl.edu/etd Recommended Citation Madsen, Erik, "Mechanisms of Copper Deficiency in the Zebrafish Embryo" (2010). All Theses and Dissertations (ETDs). 220. https://openscholarship.wustl.edu/etd/220 This Dissertation is brought to you for free and open access by Washington University Open Scholarship. It has been accepted for inclusion in All Theses and Dissertations (ETDs) by an authorized administrator of Washington University Open Scholarship. For more information, please contact [email protected]. WASHINGTON UNIVERSITY Division of Biology and Biomedical Sciences Molecular Cell Biology Dissertation Examination Committee Jonathan Gitlin, Chair John Atkinson Guojun Bu Aaron DiAntonio Steven Johnson Jeanne Nerbonne David Wilson MECHANISMS OF COPPER DEFICIENCY IN THE ZEBRAFISH EMBRYO by Erik Christian Madsen A dissertation presented to the Graduate School of Arts and Sciences of Washington University in partial fulfillment of the requirements for the degree of Doctor of Philosophy May 2010 Saint Louis, Missouri ABSTRACT OF THE DISSERTATION Mechanisms of Copper Deficiency in the Zebrafish Embryo By Erik Christian Madsen Doctor of Philosophy in Biology and Biomedical Sciences (Molecular Cell Biology) Washington University in St. Louis, 2010 Professor Jonathan D. Gitlin, Chairperson Proper maternal nutrition is critical for early embryonic development. Despite overwhelming epidemiologic data indicating the benefits nutrient supplementation for the developing organism we do not fully understand the genetics of predisposition to abnormal developmental phenotypes when faced with suboptimal nutrient levels. -

25. C:\Documents and Settings\Kwang-Il\My
The human disease network Goh K-I, Cusick ME, Valle D, Childs B, Vidal M, Barabasi′ A-L (2007) Proc Natl Acad Sci USA 104:8685-8690 Disorder Class Bone Coats Cancer Urolithiasise Osteopetrosis disease NDP Caffey van_Buchem Exudative Cardiovascular disease disease vitreoretinopathy Norrie SLC34A1 disease 439 LRP5 Connective tissue disorder Nevo Hyperostosis, syndrome COL1A1 endosteal Dermatological PLOD1 217 PAX9 Oligodontia Osteogenesis Osteoporosis 1164 Developmental Ehlers-Danlos imperfecta syndrome Arthropathy COL3A1 Hypodontia Ear, Nose, Throat Aneurysm, COL1A2 familial_arterial Myasthenic Witkop 733 syndrome Heart syndrome Pseudoachondroplasia Endocrine 3-methylglutaconicaciduria OPA3 WISP3 Optic Marfan block MSX1 atrophy OPA1 Aortic syndrome Paramyotonia Sick_sinus Gastrointestinal aneurysm congenita syndrome 3558 Intervertebral_disc Brugada SCN4A disease syndrome Syndactyly Spondyloepiphyseal COMP COL9A2 Hematological Glaucoma Weill-Marchesani Shprintzen-Goldberg Cramps, SCN5A Zlotogora-Ogur Cleft dysplasia syndrome syndrome potassium-aggravated Myotonia 2785 syndrome palate Parkes_Weber Basal_cell FBN1 congenita Oculodentodigital COL9A3 1432 Immunological 1414 CYP1B1 syndrome nevus_syndrome MASS Hypokalemic Acquired dysplasia Peters long_QT_syndrome Epiphyseal FLNB RASA1 PTCH Keratitis syndrome periodic MATN3 Metabolic SHH anomaly Eye Ectopia Thyrotoxic paralysis dysplasia Atelosteogenesis anomalies Marshall Larson Capillary Basal_cell Holoprosencephaly Coloboma, periodic KCNH2 PVRL1 malformations GJA1 Incontinentia syndrome SLC26A2 -

For Hemochromatosis (Atransferrinemia/Hemosiderosis/Iron Absorption) C
Proc. Natl. Acad. Sci. USA Vol. 84, pp. 3457-3461, May 1987 Medical Sciences Tissue distribution and clearance kinetics of non-transferrin-bound iron in the hypotransferrinemic mouse: A rodent model for hemochromatosis (atransferrinemia/hemosiderosis/iron absorption) C. M. CRAVEN*, J. ALEXANDERt, M. ELDRIDGE*, J. P. KUSHNERt, S. BERNSTEINt, AND J. KAPLAN*§ Departments of *Pathology and tMedicine, University of Utah College of Medicine, Salt Lake City, UT 84132; and tThe Jackson Laboratories, Bar Harbor, ME 04609 Communicated by Gilbert Ashwell, January 29, 1987 (receivedfor review October 28, 1986) ABSTRACT Genetically hypotransferrinemic mice accumu- (S.B., unpublished data; see refs. 9-11). These mice (HP) late iron in the liver and pancreas. A similar pattern oftissue iron were found to have a hypochromic microcytic anemia and accumulation occurs in humans with hereditary hemohroma- were growth-retarded at birth. Neonatal HP homozygotes die tosis. In both disorders, there is a decreased plasma concentration soon after birth but can be life-spared by weekly injections of of apotransferrin. To test the hypothesis that nontransferrin- whole mouse serum or Tf, even though the serum Tf levels bound iron exists and is cleared by the parenchymal tissues, the in the life-spared animals rarely exceeded 1% of normal tissue distribution of "9Fe was studied in animals lacking values. The life-spared adults develop parenchymal iron apotransferrin. Two groups of animal were used: normal rats overload with massive iron deposition in the liver and and mice whose transferrin had been saturated by an intravenous pancreas. The parenchymal iron accumulation was not injection of nonradiolabeled iron, and mice with congenital caused simply by the serum injections, as increased tissue hypotransferrinemia. -

The Postnatal Hypotransferrinemia of Early Preterm Newborn Infants
Pediat. Res. 10: 1 18-120 (1976) Bacteriostasis newborn infection serum iron transferrin The Postnatal Hypotransferrinemia of Early Preterm Newborn Infants SAMUEL GALET AND HERBERT M. SCHULMAN'Z6' Lady Davis Institute for Medical Research, Jewish General Hospital and Biology Department, McGill University, Montreal, Quebec, Canada HARRY BARD Perinatal Service, Centre de Recherche, Hospital Ste. Justine, and Department de Pediatrie, Universire de Montreal, Montreal, Quebec, Canada Extract ble to infection, the aim of this study was to examine the transferrin levels in the sera of these infants during their first few Preterm newborns were found to be markedly hypotransfer- months of life. rinemic when compared with normal term infants. At birth the concentration of transferrin in sera from preterm infants of gestational age equal to or less than 32 weeks is 45% of that found MATERIALS AND METHODS in normal term infant sera. The preterm infant transferrin levels slowly rise so that 7-8 weeks after birth they are 78% of the level Transferrin was purified from a pool of outdated human blood found in the sera of normal term infants. We also found that the se- by a modification of a method described previously (16). Eight rum transferrin concentrations at birth correlate with gestational hundred milliliters of plasma were diluted in the cold with 800 ml age. Therefore, the transferrin levels postnatally in early preterm 0.01 M Tris-HCI, pH 8.8. Nonglobulin proteins were precipitated infants reflect postconceptional rather than postnatal age. by the addition of 1,600 ml of 0.6% Rivanol in 0.01 M Tris-HCI, pH 8.8. -

Download CGT Exome V2.0
CGT Exome version 2. -

Iron Metabolism and Iron Disorders Revisited in the Hepcidin
CENTENARY REVIEW ARTICLE Iron metabolism and iron disorders revisited Ferrata Storti Foundation in the hepcidin era Clara Camaschella,1 Antonella Nai1,2 and Laura Silvestri1,2 1Regulation of Iron Metabolism Unit, Division of Genetics and Cell Biology, San Raffaele Scientific Institute, Milan and 2Vita Salute San Raffaele University, Milan, Italy ABSTRACT Haematologica 2020 Volume 105(2):260-272 ron is biologically essential, but also potentially toxic; as such it is tightly controlled at cell and systemic levels to prevent both deficien- Icy and overload. Iron regulatory proteins post-transcriptionally con- trol genes encoding proteins that modulate iron uptake, recycling and storage and are themselves regulated by iron. The master regulator of systemic iron homeostasis is the liver peptide hepcidin, which controls serum iron through degradation of ferroportin in iron-absorptive entero- cytes and iron-recycling macrophages. This review emphasizes the most recent findings in iron biology, deregulation of the hepcidin-ferroportin axis in iron disorders and how research results have an impact on clinical disorders. Insufficient hepcidin production is central to iron overload while hepcidin excess leads to iron restriction. Mutations of hemochro- matosis genes result in iron excess by downregulating the liver BMP- SMAD signaling pathway or by causing hepcidin-resistance. In iron- loading anemias, such as β-thalassemia, enhanced albeit ineffective ery- thropoiesis releases erythroferrone, which sequesters BMP receptor lig- ands, thereby inhibiting hepcidin. In iron-refractory, iron-deficiency ane- mia mutations of the hepcidin inhibitor TMPRSS6 upregulate the BMP- Correspondence: SMAD pathway. Interleukin-6 in acute and chronic inflammation increases hepcidin levels, causing iron-restricted erythropoiesis and ane- CLARA CAMASCHELLA [email protected] mia of inflammation in the presence of iron-replete macrophages. -

H63D Syndrome in Carriers of a Homozygous Mutation of HFE Gene H63D
Incidence of a clinically relevant H63D syndrome in carriers of a homozygous mutation of HFE gene H63D Carolina Diamandis ( [email protected] ) Lazar Clinic Group David Seideman Lazar Clinic Group Jacob S. Adams H63D Syndrome Research Consortium Riku Honda H63D Syndrome Research Consortium Marianne Kaufmann H63D Syndrome Research Consortium Research Article Keywords: H63D Syndrome Posted Date: May 3rd, 2021 DOI: https://doi.org/10.21203/rs.3.rs-487488/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Incidence of a clinically relevant H63D syndrome in carriers of a homozygous mutation of HFE gene H63D Jacob S. AdamsIC, Marianne KaufmannIC, Riku HondaIC, David SeidemanLCG, Carolina DiamandisLCG Affiliations: Lazar Clinic Group (LCG) Rare Diseases Research Consortium (non-profit) International H63D Consortium (IC) (non-profit) Corresponding Author: Dr. Carolina Diamandis LCG Greece Rare Diseases Research Consortium Kifissias 16, Athina, 115 26 Hellenic Republic [email protected] ______________________________ Abstract H63D syndrome is a phenotype of a homozygous mutation of the HFE gene H63D, which is otherwise known to cause at most mild classical hemochromatosis. H63D syndrome leads to an iron overload in the body (especially in the brain, heart, liver, skin and male gonads) in the form of non-transferrin bound iron (NTBI) poisoning. Hallmark symptoms and causal factor for H63D syndrome is a mild hypotransferrinemia with transferrin saturation values >50%. H63D syndrome is an incurable multi-organ disease, leading to permanent disability. Our objective was to find out how many carriers of a homozygous H63D mutation develop H63D syndrome. For this purpose, we systematically evaluated the medical records of homozygous carriers of the mutation. -
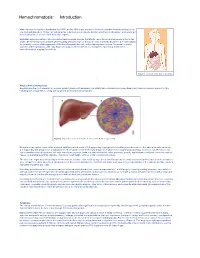
Hemochromatosis: Introduction
Hemochromatosis: Introduction Hemochromatosis was first identified in the 1800s, and by 1935 it was understood to be an inherited disease resulting in iron overload and deposition. Today, hemochromatosis is defined as a metabolic disorder affecting iron absorption , and resulting in the accumulation of excess iron in the body’s organs. Hereditary hemochromatosis (HH) is an autosomal recessive disorder that affects one in three hundred people in the United States. One in nine people carry the gene —making hemochromatosis the most common genetic disorder in the United States. It is primarily seen in middle-aged men of Northern European descent, and postmenopausal women. Treatment is readily available and the prognosis , with early diagnosis and proactive treatment, is a normal life expectancy. If untreated, hemochromatosis may lead to cirrhosis . Figure 1. Location of the liver in the body. What is Hemochromatosis? Despite being the most prevalent monoallelic genetic disease in Caucasians, hereditary hemochromatosis is under-diagnosed. There are several reasons for this, including lack of awareness, a long latency period, and nonspecific symptoms. Figure 2. Deposition of iron in the liver; A, Gross liver; B, Histological view. Normal iron absorption occurs in the proximal small intestine at a rate of 1-2 mg per day. In people with hereditary hemochromatosis, this absorption rate can reach 4–5 mg per day with progressive accumulation to 15–40 grams of iron in the body (Figure 2). Humans have no physiologic pathway to excrete iron. Therefore, iron can accumulate in any body tissue, although depositions are most common in the liver, thyroid, heart, pancreas, gonads, hypothalamus and joints. -

Download Slides (Pdf)
Iron Overload Disorders Qian Sun Clinical Chemistry Fellow National Institutes of Health DOI: 10.15428/CCTC.2018.296509 © Clinical Chemistry Outline • Iron metabolism and common iron overload disorders • Clinical presentation of iron overload disorders • Diagnostic tests • Treatment 2 Iron Metabolism Iron stored in ferritin Fe 1-2 mg/day Duodenal enterocyte Reticuloendothelial macrophage Fe Iron bound to transferrin Iron stored in ferritin Fe Synthesis of heme Liver Red cells 3 Iron Overload Disorders • Hereditary hemochromatosis (HH) • Disorders of erythroid maturation • Defects of iron transport 4 Hereditary Hemochromatosis HFE Hepcidin Fe Reticuloendothelial macrophage Ferroportin Iron release into circulation Duodenal enterocyte Fe Ferroportin Iron absorption 5 Hereditary Hemochromatosis HFE Hepcidin Fe Reticuloendothelial macrophage Ferroportin Iron release into circulation Duodenal enterocyte Fe Ferroportin Iron absorption 6 Hereditary Hemochromatosis • Most common form: HFE-associated hereditary hemochromatosis • H: hemochromatosis • Fe: Iron • 80-90% of patients are C282Y homozygotes • Prevalence of C282Y homozygosity • 1 in 200 persons of northern European ancestry • Only a small portion of C282Y homozygous subjects develop clinical disease. • Other forms of hemochromatosis: • Mutations in transferrin receptor 2 (TFR2) • Mutations in ferroportin • Juvenile hemochromatosis 7 Disorders of Erythroid Maturation • Thalassemias, congenital sideroblastic anemias, aplastic anemias • Secondary iron overload • Reduced utilization of iron