CD166 Antigen Macrophage Inflammatory Protein 3Β Dipeptidyl Peptidase 4 Clusterin Growth Regulated Oncogene Alpha C-X-C Motif C
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

METACYC ID Description A0AR23 GO:0004842 (Ubiquitin-Protein Ligase
Electronic Supplementary Material (ESI) for Integrative Biology This journal is © The Royal Society of Chemistry 2012 Heat Stress Responsive Zostera marina Genes, Southern Population (α=0. -

Identification of BIRC6 As a Novel Intervention Target For
Lamers et al. BMC Cancer 2012, 12:285 http://www.biomedcentral.com/1471-2407/12/285 RESEARCH ARTICLE Open Access Identification of BIRC6 as a novel intervention target for neuroblastoma therapy Fieke Lamers1, Linda Schild1, Jan Koster1, Frank Speleman2, Ingrid ra3, Ellen M Westerhout1, Peter van Sluis1, Rogier Versteeg1, Huib N Caron4 and Jan J Molenaar1,5* Abstract Background: Neuroblastoma are pediatric tumors of the sympathetic nervous system with a poor prognosis. Apoptosis is often deregulated in cancer cells, but only a few defects in apoptotic routes have been identified in neuroblastoma. Methods: Here we investigated genomic aberrations affecting genes of the intrinsic apoptotic pathway in neuroblastoma. We analyzed DNA profiling data (CGH and SNP arrays) and mRNA expression data of 31 genes of the intrinsic apoptotic pathway in a dataset of 88 neuroblastoma tumors using the R2 bioinformatic platform (http://r2.amc.nl). BIRC6 was selected for further analysis as a tumor driving gene. Knockdown experiments were performed using BIRC6 lentiviral shRNA and phenotype responses were analyzed by Western blot and MTT-assays. In addition, DIABLO levels and interactions were investigated with immunofluorescence and co-immunoprecipitation. Results: We observed frequent gain of the BIRC6 gene on chromosome 2, which resulted in increased mRNA expression. BIRC6 is an inhibitor of apoptosis protein (IAP), that can bind and degrade the cytoplasmic fraction of the pro-apoptotic protein DIABLO. DIABLO mRNA expression was exceptionally high in neuroblastoma but the protein was only detected in the mitochondria. Upon silencing of BIRC6 by shRNA, DIABLO protein levels increased and cells went into apoptosis. Co-immunoprecipitation confirmed direct interaction between DIABLO and BIRC6 in neuroblastoma cell lines. -

Gentaur Products List
Chapter 2 : Gentaur Products List • Rabbit Anti LAMR1 Polyclonal Antibody Cy5 Conjugated Conjugated • Rabbit Anti Podoplanin gp36 Polyclonal Antibody Cy5 • Rabbit Anti LAMR1 CT Polyclonal Antibody Cy5 • Rabbit Anti phospho NFKB p65 Ser536 Polyclonal Conjugated Conjugated Antibody Cy5 Conjugated • Rabbit Anti CHRNA7 Polyclonal Antibody Cy5 Conjugated • Rat Anti IAA Monoclonal Antibody Cy5 Conjugated • Rabbit Anti EV71 VP1 CT Polyclonal Antibody Cy5 • Rabbit Anti Connexin 40 Polyclonal Antibody Cy5 • Rabbit Anti IAA Indole 3 Acetic Acid Polyclonal Antibody Conjugated Conjugated Cy5 Conjugated • Rabbit Anti LHR CGR Polyclonal Antibody Cy5 Conjugated • Rabbit Anti Integrin beta 7 Polyclonal Antibody Cy5 • Rabbit Anti Natrexone Polyclonal Antibody Cy5 Conjugated • Rabbit Anti MMP 20 Polyclonal Antibody Cy5 Conjugated Conjugated • Rabbit Anti Melamine Polyclonal Antibody Cy5 Conjugated • Rabbit Anti BCHE NT Polyclonal Antibody Cy5 Conjugated • Rabbit Anti NAP1 NAP1L1 Polyclonal Antibody Cy5 • Rabbit Anti Acetyl p53 K382 Polyclonal Antibody Cy5 • Rabbit Anti BCHE CT Polyclonal Antibody Cy5 Conjugated Conjugated Conjugated • Rabbit Anti HPV16 E6 Polyclonal Antibody Cy5 Conjugated • Rabbit Anti CCP Polyclonal Antibody Cy5 Conjugated • Rabbit Anti JAK2 Polyclonal Antibody Cy5 Conjugated • Rabbit Anti HPV18 E6 Polyclonal Antibody Cy5 Conjugated • Rabbit Anti HDC Polyclonal Antibody Cy5 Conjugated • Rabbit Anti Microsporidia protien Polyclonal Antibody Cy5 • Rabbit Anti HPV16 E7 Polyclonal Antibody Cy5 Conjugated • Rabbit Anti Neurocan Polyclonal -

Table S1 the Four Gene Sets Derived from Gene Expression Profiles of Escs and Differentiated Cells
Table S1 The four gene sets derived from gene expression profiles of ESCs and differentiated cells Uniform High Uniform Low ES Up ES Down EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol 269261 Rpl12 11354 Abpa 68239 Krt42 15132 Hbb-bh1 67891 Rpl4 11537 Cfd 26380 Esrrb 15126 Hba-x 55949 Eef1b2 11698 Ambn 73703 Dppa2 15111 Hand2 18148 Npm1 11730 Ang3 67374 Jam2 65255 Asb4 67427 Rps20 11731 Ang2 22702 Zfp42 17292 Mesp1 15481 Hspa8 11807 Apoa2 58865 Tdh 19737 Rgs5 100041686 LOC100041686 11814 Apoc3 26388 Ifi202b 225518 Prdm6 11983 Atpif1 11945 Atp4b 11614 Nr0b1 20378 Frzb 19241 Tmsb4x 12007 Azgp1 76815 Calcoco2 12767 Cxcr4 20116 Rps8 12044 Bcl2a1a 219132 D14Ertd668e 103889 Hoxb2 20103 Rps5 12047 Bcl2a1d 381411 Gm1967 17701 Msx1 14694 Gnb2l1 12049 Bcl2l10 20899 Stra8 23796 Aplnr 19941 Rpl26 12096 Bglap1 78625 1700061G19Rik 12627 Cfc1 12070 Ngfrap1 12097 Bglap2 21816 Tgm1 12622 Cer1 19989 Rpl7 12267 C3ar1 67405 Nts 21385 Tbx2 19896 Rpl10a 12279 C9 435337 EG435337 56720 Tdo2 20044 Rps14 12391 Cav3 545913 Zscan4d 16869 Lhx1 19175 Psmb6 12409 Cbr2 244448 Triml1 22253 Unc5c 22627 Ywhae 12477 Ctla4 69134 2200001I15Rik 14174 Fgf3 19951 Rpl32 12523 Cd84 66065 Hsd17b14 16542 Kdr 66152 1110020P15Rik 12524 Cd86 81879 Tcfcp2l1 15122 Hba-a1 66489 Rpl35 12640 Cga 17907 Mylpf 15414 Hoxb6 15519 Hsp90aa1 12642 Ch25h 26424 Nr5a2 210530 Leprel1 66483 Rpl36al 12655 Chi3l3 83560 Tex14 12338 Capn6 27370 Rps26 12796 Camp 17450 Morc1 20671 Sox17 66576 Uqcrh 12869 Cox8b 79455 Pdcl2 20613 Snai1 22154 Tubb5 12959 Cryba4 231821 Centa1 17897 -

Diplomarbeit
DIPLOMARBEIT Titel der Diplomarbeit „The influence of free fatty acids on the development of liver inflammation“ Verfasser Mario Kuttke, B.Sc. angestrebter akademischer Grad Magister der Naturwissenschaften (Mag.rer.nat.) Wien, 2012 Studienkennzahl lt. Studienblatt: A 490 Studienrichtung lt. Studienblatt: Diplomstudium Molekulare Biologie Betreuerin / Betreuer: A.o.Univ.-Prof.Dipl.-Ing.Dr. Marcela Hermann Danksagung Zuerst möchte ich mich bei a.o.Univ.-Prof. Dipl.-Ing. Dr. Marcela Hermann für die Betreuung meiner Diplomarbeit bedanken. Besonderer Dank gilt a.o.Univ-Prof. Dr. Bettina Grasl-Kraupp für die fachliche Unterstützung und Betreuung während der praktischen Durchführung der Arbeit. Weiters bedanke ich mich bei Sandra Sagmeister, Therese Böhm, Nora Bintner, Waltraud Schrottmaier, Melanie Pichlbauer, Marzieh Nejabat, Teresa Riegler, Bettina Wingelhofer und Christiane Maier für die ausgezeichnete Zusammenarbeit im Labor und die Unterstützung in allen Lebenslagen. Birgit Mir-Karner, Helga Koudelka und Krystyna Bukowska danke ich für ihre Hilfsbereitschaft und für die kollegiale Zusammenarbeit. Mein größter Dank gilt meinen Eltern, Ursula und Heinz, und meiner Großmutter, Theresia, die mir mein Studium ermöglicht und mich immer unterstützt haben, sowie meinem Bruder, Alex, der in allen Lebenslagen für mich da ist. Table of Contents TABLE OF CONTENTS INTRODUCTION ............................................................................................................................................. 4 HEPATOCELLULAR CARCINOMA (HCC) -

Exploring the Metastatic Role of the Inhibitor of Apoptosis BIRC6 in Breast Cancer
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.08.438518; this version posted April 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Exploring the metastatic role of the inhibitor of apoptosis BIRC6 in Breast 2 Cancer 3 Corresponding author: Matias Luis Pidre, Pringles 3010, Lanús, Buenos Aires, Argentina, CP 1824 4 [email protected], mobile: +54 9 221 364 6836 5 AUTHORS 6 Santiago M. Gómez Bergna1; Abril Marchesini1; Leslie C. Amorós Morales1; Paula N. Arrías1; Hernán 7 G. Farina2; Víctor Romanowski1; M. Florencia Gottardo2*; Matias L. Pidre1*. 8 *Both authors equally contributed to this work. 9 AUTHOR AFFILIATIONS 10 1Instituto de Biotecnología y biología molecular (IBBM-CONICET-UNLP) 11 2Center of Molecular & Translational Oncology, Department of Science and Technology, 12 National University of Quilmes, Buenos Aires, Argentina. 13 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.04.08.438518; this version posted April 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 14 Abstract 15 Breast cancer is the most common cancer as well as the first cause of death by cancer in 16 women worldwide. BIRC6 (baculoviral IAP repeat-containing protein 6) is a member of the 17 inhibitors of apoptosis protein family thought to play an important role in the progression or 18 chemoresistance of many cancers. The aim of the present work was to investigate the role of 19 apoptosis inhibitor BIRC6 in breast cancer, focusing particularly on its involvement in the 20 metastatic cascade. -

Summary & Conclusions
Enzymatic functionalization and degradation of natural and synthetic polymers Von der Fakultät für Mathematik, Informatik und Naturwissenschaften der RWTH Aachen University zur Erlangung des akademischen Grades einer Doktorin der Naturwissenschaften genehmigte Dissertation vorgelegt von Shohana Subrin Islam M. Sc. Biotechnologie aus Dhaka, Bangladesch Berichter: Univ. -Prof. Dr. Ulrich Schwaneberg Univ. -Prof. Dr. Lothar Elling Tag der mündlichen Prüfung: 23.01.2019 Diese Dissertation ist auf den Internetseiten der Universitätsbibliothek verfügbar. To my mom & my sister-the two persons in the world who always stand by me Table of content Table of content Table of content _______________________________________________________________ v Publications and patents ________________________________________________________ ix Abstract _____________________________________________________________________ xi 1. General introduction _______________________________________________________ 1 1.1 Enzymatic functionalization of (bio)polymers _______________________________________ 1 1.2 Enzymatic degradation of polymers _______________________________________________ 3 1.3 Protein engineering ____________________________________________________________ 5 1.3.1 Directed evolution of enzymes _________________________________________________________ 6 1.3.2 KnowVolution – Directed Evolution 2.0 __________________________________________________ 9 1.4 Aims of the dissertation _______________________________________________________ 11 2. Engineering of -

Phylogenetic Analysis, Subcellular Localization, and Expression
BMC Plant Biology BioMed Central Research article Open Access Phylogenetic analysis, subcellular localization, and expression patterns of RPD3/HDA1 family histone deacetylases in plants Malona V Alinsug, Chun-Wei Yu and Keqiang Wu* Address: Institute of Plant Biology, College of Life Science, National Taiwan University, Taipei, Taiwan Email: Malona V Alinsug - [email protected]; Chun-Wei Yu - [email protected]; Keqiang Wu* - [email protected] * Corresponding author Published: 28 March 2009 Received: 26 November 2008 Accepted: 28 March 2009 BMC Plant Biology 2009, 9:37 doi:10.1186/1471-2229-9-37 This article is available from: http://www.biomedcentral.com/1471-2229/9/37 © 2009 Alinsug et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: Although histone deacetylases from model organisms have been previously identified, there is no clear basis for the classification of histone deacetylases under the RPD3/ HDA1 superfamily, particularly on plants. Thus, this study aims to reconstruct a phylogenetic tree to determine evolutionary relationships between RPD3/HDA1 histone deacetylases from six different plants representing dicots with Arabidopsis thaliana, Populus trichocarpa, and Pinus taeda, monocots with Oryza sativa and Zea mays, and the lower plants with Physcomitrella patens. Results: Sixty two histone deacetylases of RPD3/HDA1 family from the six plant species were phylogenetically analyzed to determine corresponding orthologues. Three clusters were formed separating Class I, Class II, and Class IV. -

MOUSE INTERLEUKIN-28B/INTERFERON-LAMBDA 3, CARRIER FREE Product Number: 12821-1 Lot Number: 5559 Size: 25 Μg
MOUSE INTERLEUKIN-28B/INTERFERON-LAMBDA 3, CARRIER FREE Product Number: 12821-1 Lot Number: 5559 Size: 25 µg Description: Mouse Interleukin-28B/Interferon-lambda 3, Carrier Free Source: A DNA sequence encoding the mature mouse IL-28B/IFN-λ3 (Asp 20 - Val 193) (Kotenko, S.V. et al., 2003, Nat. Immunol. 4(1):69 - 77) was expressed in E. coli. Form: Lyophilized Buffer: Phosphate-buffered saline (PBS) Reconstitution: It is recommended that sterile PBS be added to the vial to prepare a stock solution of no less than 100 μg/mL. Endotoxin: < 1 EU/µg Molecular Weight: The 175 amino acid residue methionyl form of recombinant mouse IL-28B has a predicted molecular mass of approximately 19.7 kDa. Purity: > 95% Synonyms: Mu IL-28B; Mu IFN-λ3 Accession #: NP_796370 Assays Used to Measure Bioactivity: Human HepG2 cells infected with encephalomyocarditis virus (Sheppard, P. et al., 2003, Nature Immunol. 4:63). The ED50 for this effect is typically 7.5 - 37.5 ng/mL. Shipping Conditions: Wet Ice Physical State of Product During Shipping: Lyophilized Special Conditions/Comments: After receipt, this product should be kept at -70˚C or below for retention of full activity. Upon reconstitution, this cytokine can be stored under sterile conditions at 2˚C to 8˚C for one month or at -20˚C to -70˚C in a manual defrost freezer for three months without detection loss of activity. Avoid repeated freeze-thaw cycles. For more information on protein handling, visit the PBL website at www.interferonsource.com . Product Information: Human IL-28A, IL-28B, and IL-29, also named interferon-λ2 (IFN-λ2), IFN-λ3, and IFN-λ1, respectively, are newly identified class II cytokine receptor ligands that are distantly related to members of the IL-10 family (11- 13% aa sequence identity) and type I IFN family (15 - 19% aa sequence identity).1 – 3 The genes encoding these three cytokines are localized to chromosome 19 and each is composed of multiple exons. -

HMGB1 in Health and Disease R
Donald and Barbara Zucker School of Medicine Journal Articles Academic Works 2014 HMGB1 in health and disease R. Kang R. C. Chen Q. H. Zhang W. Hou S. Wu See next page for additional authors Follow this and additional works at: https://academicworks.medicine.hofstra.edu/articles Part of the Emergency Medicine Commons Recommended Citation Kang R, Chen R, Zhang Q, Hou W, Wu S, Fan X, Yan Z, Sun X, Wang H, Tang D, . HMGB1 in health and disease. 2014 Jan 01; 40():Article 533 [ p.]. Available from: https://academicworks.medicine.hofstra.edu/articles/533. Free full text article. This Article is brought to you for free and open access by Donald and Barbara Zucker School of Medicine Academic Works. It has been accepted for inclusion in Journal Articles by an authorized administrator of Donald and Barbara Zucker School of Medicine Academic Works. Authors R. Kang, R. C. Chen, Q. H. Zhang, W. Hou, S. Wu, X. G. Fan, Z. W. Yan, X. F. Sun, H. C. Wang, D. L. Tang, and +8 additional authors This article is available at Donald and Barbara Zucker School of Medicine Academic Works: https://academicworks.medicine.hofstra.edu/articles/533 NIH Public Access Author Manuscript Mol Aspects Med. Author manuscript; available in PMC 2015 December 01. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Mol Aspects Med. 2014 December ; 0: 1–116. doi:10.1016/j.mam.2014.05.001. HMGB1 in Health and Disease Rui Kang1,*, Ruochan Chen1, Qiuhong Zhang1, Wen Hou1, Sha Wu1, Lizhi Cao2, Jin Huang3, Yan Yu2, Xue-gong Fan4, Zhengwen Yan1,5, Xiaofang Sun6, Haichao Wang7, Qingde Wang1, Allan Tsung1, Timothy R. -

Abstracts from the 10Th World Congress for Microcirculation
DOI:10.1111/micc.12246 Abstracts Abstracts from the 10th World Congress for Microcirculation September 25th – 27th, 2015 Kyoto, Japan PLENARY LECTURES vascular neutrophils that constantly patrol the lung and can detect and phagocytose bacteria attached to endothelium suggesting some communication inter-cellular communica- PL1 tion. Clearly much anti-microbial activity occurs in the Imaging the microcirculation in infections vasculature prior to dissemination of bacteria into tissues. P Kubes University of Calgary, Calgary, Canada PL2 Using spinning disk microscopy has allowed us to track pathogens and immune cells in the microcirculation. Mapping oxygen in the brain of awake resting Observing the liver microvasculature revealed numerous mice immune speed bumps that slowed, delayed and even S Charpak prevented bacteria from disseminating to other organs. For Laboratory of Neurophysiology and New Microscopies, Inserm U1128, example, when Borrelia burgdorferi enters the vasculature Paris Descartes University, Paris, France they are immediately caught by the liver vascular macro- The brain is extremely sensitive to hypoxia. Yet, the phage, the Kupffer cells and are engulfed and antigens are physiological values of oxygen concentration in the brain presented on CD1d to resident vascular immune cells remain elusive because high resolution measurements have including invariant Natural Killer T cells (iNKT cells). These only been performed during anesthesia, which affects two iNKT cells receive messages and rapidly make gamma main parameters modulating tissue oxygenation, i.e. interferon to help with immunity. Absence of these events neuronal activity and cerebral blood flow. Using the recent leads to massive dissemination of borrelia especially to joints. finding that measurements of capillary erythrocyte-associ- In the joints the iNKT cells live closely apposed but outside ated transients i.e. -
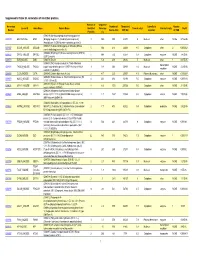
Supporting Table S3 for PDF Maker
Supplemental Table S3. Annotation of identified proteins. Number of Sequence Accession Number of Theoretical Subcellular Number Locus ID Gene Name Protein Name Identified Coverage Theoretical pI Protein Family NSAF Number Amino Acid MW (Da) Location of TMD Peptides (%) (O08539) Myc box-dependent-interacting protein 1 O08539 BIN1_MOUSE BIN1 (Bridging integrator 1) (Amphiphysin-like protein) 3 10.5 588 64470 5 Nucleus other NONE 5.73E-05 (Amphiphysin II) (SH3-domain-containing protein 9) (O08547) Vesicle-trafficking protein SEC22b (SEC22 O08547 SC22B_MOUSE SEC22B 5 30.4 214 24609 8.5 Cytoplasm other 2 0.000262 vesicle-trafficking protein-like 1) (O08553) Dihydropyrimidinase-related protein 2 (DRP-2) O08553 DPYL2_MOUSE DPYSL2 5 19.4 572 62278 6.4 Cytoplasm enzyme NONE 7.85E-05 (ULIP 2 protein) O08579 EMD_MOUSE EMD (O08579) Emerin 1 5.8 259 29436 5 Nucleus other 1 8.67E-05 (O08583) THO complex subunit 4 (Tho4) (RNA and transcription O08583 THOC4_MOUSE THOC4 export factor-binding protein 1) (REF1-I) (Ally of AML-1 1 9.8 254 26809 11.2 Nucleus NONE 2.21E-05 regulator and LEF-1) (Aly/REF) O08585 CLCA_MOUSE CLTA (O08585) Clathrin light chain A (Lca) 2 4.7 235 25557 4.5 Plasma Membrane other NONE 0.000287 (O08600) Endonuclease G, mitochondrial precursor (EC O08600 NUCG_MOUSE ENDOG 4 23.1 294 32191 9.5 Cytoplasm enzyme NONE 0.000134 3.1.30.-) (Endo G) (O08638) Myosin-11 (Myosin heavy chain, smooth O08638 MYH11_MOUSE MYH11 8 6.6 1972 227026 5.5 Cytoplasm other NONE 3.13E-05 muscle isoform) (SMMHC) (O08648) Mitogen-activated protein kinase kinase