Animal Phylogeny and Its Evolutionary Implications∗
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Systema Naturae∗
Systema Naturae∗ c Alexey B. Shipunov v. 5.802 (June 29, 2008) 7 Regnum Monera [ Bacillus ] /Bacteria Subregnum Bacteria [ 6:8Bacillus ]1 Superphylum Posibacteria [ 6:2Bacillus ] stat.m. Phylum 1. Firmicutes [ 6Bacillus ]2 Classis 1(1). Thermotogae [ 5Thermotoga ] i.s. 2(2). Mollicutes [ 5Mycoplasma ] 3(3). Clostridia [ 5Clostridium ]3 4(4). Bacilli [ 5Bacillus ] 5(5). Symbiobacteres [ 5Symbiobacterium ] Phylum 2. Actinobacteria [ 6Actynomyces ] Classis 1(6). Actinobacteres [ 5Actinomyces ] Phylum 3. Hadobacteria [ 6Deinococcus ] sed.m. Classis 1(7). Hadobacteres [ 5Deinococcus ]4 Superphylum Negibacteria [ 6:2Rhodospirillum ] stat.m. Phylum 4. Chlorobacteria [ 6Chloroflexus ]5 Classis 1(8). Ktedonobacteres [ 5Ktedonobacter ] sed.m. 2(9). Thermomicrobia [ 5Thermomicrobium ] 3(10). Chloroflexi [ 5Chloroflexus ] ∗Only recent taxa. Viruses are not included. Abbreviations and signs: sed.m. (sedis mutabilis); stat.m. (status mutabilis): s., aut i. (superior, aut interior); i.s. (incertae sedis); sed.p. (sedis possibilis); s.str. (sensu stricto); s.l. (sensu lato); incl. (inclusum); excl. (exclusum); \quotes" for environmental groups; * (asterisk) for paraphyletic taxa; / (slash) at margins for major clades (\domains"). 1Incl. \Nanobacteria" i.s. et dubitativa, \OP11 group" i.s. 2Incl. \TM7" i.s., \OP9", \OP10". 3Incl. Dictyoglomi sed.m., Fusobacteria, Thermolithobacteria. 4= Deinococcus{Thermus. 5Incl. Thermobaculum i.s. 1 4(11). Dehalococcoidetes [ 5Dehalococcoides ] 5(12). Anaerolineae [ 5Anaerolinea ]6 Phylum 5. Cyanobacteria [ 6Nostoc ] Classis 1(13). Gloeobacteres [ 5Gloeobacter ] 2(14). Chroobacteres [ 5Chroococcus ]7 3(15). Hormogoneae [ 5Nostoc ] Phylum 6. Bacteroidobacteria [ 6Bacteroides ]8 Classis 1(16). Fibrobacteres [ 5Fibrobacter ] 2(17). Chlorobi [ 5Chlorobium ] 3(18). Salinibacteres [ 5Salinibacter ] 4(19). Bacteroidetes [ 5Bacteroides ]9 Phylum 7. Spirobacteria [ 6Spirochaeta ] Classis 1(20). Spirochaetes [ 5Spirochaeta ] s.l.10 Phylum 8. Planctobacteria [ 6Planctomyces ]11 Classis 1(21). -

Identical Genomic Organization of Two Hemichordate Hox Clusters
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Current Biology 22, 2053–2058, November 6, 2012 ª2012 Elsevier Ltd All rights reserved http://dx.doi.org/10.1016/j.cub.2012.08.052 Report Identical Genomic Organization of Two Hemichordate Hox Clusters Robert Freeman,1,12 Tetsuro Ikuta,2,12 Michael Wu,3 [10] have similar organization to the hemichordate cluster, Ryo Koyanagi,2 Takeshi Kawashima,2 Kunifumi Tagawa,4 but with different posterior genes. These results provide Tom Humphreys,5 Guang-Chen Fang,6 Asao Fujiyama,7 genomic evidence for a well-ordered complex in the deutero- Hidetoshi Saiga,8 Christopher Lowe,9 Kim Worley,10 stome ancestor for the hox1–hox9/10 region, with the Jerry Jenkins,11 Jeremy Schmutz,11 Marc Kirschner,1 number and kind of posterior genes still to be elucidated. Daniel Rokhsar,3 Nori Satoh,2,* and John Gerhart3,* 1 Department of Systems Biology, Harvard Medical School, Results and Discussion Boston, MA 02114, USA 2 Marine Genomics Unit, Okinawa Institute of Science and Here we characterize the order, transcriptional orientation, and Technology Graduate University, Onna, clustering of the Hox genes of the genomes of two widely Okinawa 904-0495, Japan studied model hemichordates, Saccoglossus kowalevskii 3 Department of Molecular and Cell Biology, University of and Ptychodera flava [4, 11, 12], that represent two major California, Berkeley, Berkeley, CA 94720-3200, USA evolutionary branches of enteropneust hemichordates that 4 Marine Biological Laboratory, Graduate School of Science, diverged an estimated 400 million years ago (MYa) [13]: Hiroshima University, Onomichi, Hiroshima 722-0073, Japan Saccoglossus, of the direct developing harimaniids, and 5 Department of Cell and Molecular Biology, University of Ptychodera, of the indirect developing ptychoderids [14]. -

Platyhelminthes, Nemertea, and "Aschelminthes" - A
BIOLOGICAL SCIENCE FUNDAMENTALS AND SYSTEMATICS – Vol. III - Platyhelminthes, Nemertea, and "Aschelminthes" - A. Schmidt-Rhaesa PLATYHELMINTHES, NEMERTEA, AND “ASCHELMINTHES” A. Schmidt-Rhaesa University of Bielefeld, Germany Keywords: Platyhelminthes, Nemertea, Gnathifera, Gnathostomulida, Micrognathozoa, Rotifera, Acanthocephala, Cycliophora, Nemathelminthes, Gastrotricha, Nematoda, Nematomorpha, Priapulida, Kinorhyncha, Loricifera Contents 1. Introduction 2. General Morphology 3. Platyhelminthes, the Flatworms 4. Nemertea (Nemertini), the Ribbon Worms 5. “Aschelminthes” 5.1. Gnathifera 5.1.1. Gnathostomulida 5.1.2. Micrognathozoa (Limnognathia maerski) 5.1.3. Rotifera 5.1.4. Acanthocephala 5.1.5. Cycliophora (Symbion pandora) 5.2. Nemathelminthes 5.2.1. Gastrotricha 5.2.2. Nematoda, the Roundworms 5.2.3. Nematomorpha, the Horsehair Worms 5.2.4. Priapulida 5.2.5. Kinorhyncha 5.2.6. Loricifera Acknowledgements Glossary Bibliography Biographical Sketch Summary UNESCO – EOLSS This chapter provides information on several basal bilaterian groups: flatworms, nemerteans, Gnathifera,SAMPLE and Nemathelminthes. CHAPTERS These include species-rich taxa such as Nematoda and Platyhelminthes, and as taxa with few or even only one species, such as Micrognathozoa (Limnognathia maerski) and Cycliophora (Symbion pandora). All Acanthocephala and subgroups of Platyhelminthes and Nematoda, are parasites that often exhibit complex life cycles. Most of the taxa described are marine, but some have also invaded freshwater or the terrestrial environment. “Aschelminthes” are not a natural group, instead, two taxa have been recognized that were earlier summarized under this name. Gnathifera include taxa with a conspicuous jaw apparatus such as Gnathostomulida, Micrognathozoa, and Rotifera. Although they do not possess a jaw apparatus, Acanthocephala also belong to Gnathifera due to their epidermal structure. ©Encyclopedia of Life Support Systems (EOLSS) BIOLOGICAL SCIENCE FUNDAMENTALS AND SYSTEMATICS – Vol. -

New Zealand's Genetic Diversity
1.13 NEW ZEALAND’S GENETIC DIVERSITY NEW ZEALAND’S GENETIC DIVERSITY Dennis P. Gordon National Institute of Water and Atmospheric Research, Private Bag 14901, Kilbirnie, Wellington 6022, New Zealand ABSTRACT: The known genetic diversity represented by the New Zealand biota is reviewed and summarised, largely based on a recently published New Zealand inventory of biodiversity. All kingdoms and eukaryote phyla are covered, updated to refl ect the latest phylogenetic view of Eukaryota. The total known biota comprises a nominal 57 406 species (c. 48 640 described). Subtraction of the 4889 naturalised-alien species gives a biota of 52 517 native species. A minimum (the status of a number of the unnamed species is uncertain) of 27 380 (52%) of these species are endemic (cf. 26% for Fungi, 38% for all marine species, 46% for marine Animalia, 68% for all Animalia, 78% for vascular plants and 91% for terrestrial Animalia). In passing, examples are given both of the roles of the major taxa in providing ecosystem services and of the use of genetic resources in the New Zealand economy. Key words: Animalia, Chromista, freshwater, Fungi, genetic diversity, marine, New Zealand, Prokaryota, Protozoa, terrestrial. INTRODUCTION Article 10b of the CBD calls for signatories to ‘Adopt The original brief for this chapter was to review New Zealand’s measures relating to the use of biological resources [i.e. genetic genetic resources. The OECD defi nition of genetic resources resources] to avoid or minimize adverse impacts on biological is ‘genetic material of plants, animals or micro-organisms of diversity [e.g. genetic diversity]’ (my parentheses). -

The Closest Unicellular Relatives of Animals
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Current Biology, Vol. 12, 1773–1778, October 15, 2002, 2002 Elsevier Science Ltd. All rights reserved. PII S0960-9822(02)01187-9 The Closest Unicellular Relatives of Animals B.F. Lang,1,2 C. O’Kelly,1,3 T. Nerad,4 M.W. Gray,1,5 Results and Discussion and G. Burger1,2,6 1The Canadian Institute for Advanced Research The evolution of the Metazoa from single-celled protists Program in Evolutionary Biology is an issue that has intrigued biologists for more than a 2 De´ partement de Biochimie century. Early morphological and more recent ultra- Universite´ de Montre´ al structural and molecular studies have converged in sup- Succursale Centre-Ville porting the now widely accepted view that animals are Montre´ al, Que´ bec H3C 3J7 related to Fungi, choanoflagellates, and ichthyosporean Canada protists. However, controversy persists as to the spe- 3 Bigelow Laboratory for Ocean Sciences cific evolutionary relationships among these major P.O. Box 475 groups. This uncertainty is reflected in the plethora of 180 McKown Point Road published molecular phylogenies that propose virtually West Boothbay Harbor, Maine 04575 all of the possible alternative tree topologies involving 4 American Type Culture Collection Choanoflagellata, Fungi, Ichthyosporea, and Metazoa. 10801 University Boulevard For example, a monophyletic MetazoaϩChoanoflagel- Manassas, Virginia 20110 lata group has been suggested on the basis of small 5 Department of Biochemistry subunit (SSU) rDNA sequences [1, 6, 7]. Other studies and Molecular Biology using the same sequences have allied Choanoflagellata Dalhousie University with the Fungi [8], placed Choanoflagellata prior to the Halifax, Nova Scotia B3H 4H7 divergence of animals and Fungi [9], or even placed Canada them prior to the divergence of green algae and land plants [10]. -

Sex Is a Ubiquitous, Ancient, and Inherent Attribute of Eukaryotic Life
PAPER Sex is a ubiquitous, ancient, and inherent attribute of COLLOQUIUM eukaryotic life Dave Speijera,1, Julius Lukešb,c, and Marek Eliášd,1 aDepartment of Medical Biochemistry, Academic Medical Center, University of Amsterdam, 1105 AZ, Amsterdam, The Netherlands; bInstitute of Parasitology, Biology Centre, Czech Academy of Sciences, and Faculty of Sciences, University of South Bohemia, 370 05 Ceské Budejovice, Czech Republic; cCanadian Institute for Advanced Research, Toronto, ON, Canada M5G 1Z8; and dDepartment of Biology and Ecology, University of Ostrava, 710 00 Ostrava, Czech Republic Edited by John C. Avise, University of California, Irvine, CA, and approved April 8, 2015 (received for review February 14, 2015) Sexual reproduction and clonality in eukaryotes are mostly Sex in Eukaryotic Microorganisms: More Voyeurs Needed seen as exclusive, the latter being rather exceptional. This view Whereas absence of sex is considered as something scandalous for might be biased by focusing almost exclusively on metazoans. a zoologist, scientists studying protists, which represent the ma- We analyze and discuss reproduction in the context of extant jority of extant eukaryotic diversity (2), are much more ready to eukaryotic diversity, paying special attention to protists. We accept that a particular eukaryotic group has not shown any evi- present results of phylogenetically extended searches for ho- dence of sexual processes. Although sex is very well documented mologs of two proteins functioning in cell and nuclear fusion, in many protist groups, and members of some taxa, such as ciliates respectively (HAP2 and GEX1), providing indirect evidence for (Alveolata), diatoms (Stramenopiles), or green algae (Chlor- these processes in several eukaryotic lineages where sex has oplastida), even serve as models to study various aspects of sex- – not been observed yet. -

Multigene Eukaryote Phylogeny Reveals the Likely Protozoan Ancestors of Opis- Thokonts (Animals, Fungi, Choanozoans) and Amoebozoa
Accepted Manuscript Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opis- thokonts (animals, fungi, choanozoans) and Amoebozoa Thomas Cavalier-Smith, Ema E. Chao, Elizabeth A. Snell, Cédric Berney, Anna Maria Fiore-Donno, Rhodri Lewis PII: S1055-7903(14)00279-6 DOI: http://dx.doi.org/10.1016/j.ympev.2014.08.012 Reference: YMPEV 4996 To appear in: Molecular Phylogenetics and Evolution Received Date: 24 January 2014 Revised Date: 2 August 2014 Accepted Date: 11 August 2014 Please cite this article as: Cavalier-Smith, T., Chao, E.E., Snell, E.A., Berney, C., Fiore-Donno, A.M., Lewis, R., Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opisthokonts (animals, fungi, choanozoans) and Amoebozoa, Molecular Phylogenetics and Evolution (2014), doi: http://dx.doi.org/10.1016/ j.ympev.2014.08.012 This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. 1 1 Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opisthokonts 2 (animals, fungi, choanozoans) and Amoebozoa 3 4 Thomas Cavalier-Smith1, Ema E. Chao1, Elizabeth A. Snell1, Cédric Berney1,2, Anna Maria 5 Fiore-Donno1,3, and Rhodri Lewis1 6 7 1Department of Zoology, University of Oxford, South Parks Road, Oxford OX1 3PS, UK. -
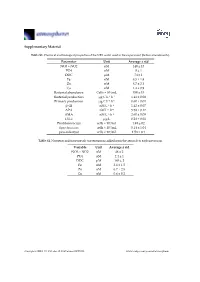
Supplementary Material Parameter Unit Average ± Std NO3 + NO2 Nm
Supplementary Material Table S1. Chemical and biological properties of the NRS water used in the experiment (before amendments). Parameter Unit Average ± std NO3 + NO2 nM 140 ± 13 PO4 nM 8 ± 1 DOC μM 74 ± 1 Fe nM 8.5 ± 1.8 Zn nM 8.7 ± 2.1 Cu nM 1.4 ± 0.9 Bacterial abundance Cells × 104/mL 350 ± 15 Bacterial production μg C L−1 h−1 1.41 ± 0.08 Primary production μg C L−1 h−1 0.60 ± 0.01 β-Gl nM L−1 h−1 1.42 ± 0.07 APA nM L−1 h−1 5.58 ± 0.17 AMA nM L−1·h−1 2.60 ± 0.09 Chl-a μg/L 0.28 ± 0.01 Prochlorococcus cells × 104/mL 1.49 ± 02 Synechococcus cells × 104/mL 5.14 ± 1.04 pico-eukaryot cells × 103/mL 1.58 × 0.1 Table S2. Nutrients and trace metals concentrations added from the aerosols to each mesocosm. Variable Unit Average ± std NO3 + NO2 nM 48 ± 2 PO4 nM 2.4 ± 1 DOC μM 165 ± 2 Fe nM 2.6 ± 1.5 Zn nM 6.7 ± 2.5 Cu nM 0.6 ± 0.2 Atmosphere 2019, 10, 358; doi:10.3390/atmos10070358 www.mdpi.com/journal/atmosphere Atmosphere 2019, 10, 358 2 of 6 Table S3. ANOVA test results between control, ‘UV-treated’ and ‘live-dust’ treatments at 20 h or 44 h, with significantly different values shown in bold. ANOVA df Sum Sq Mean Sq F Value p-value Chl-a 20 H 2, 6 0.03, 0.02 0.02, 0 4.52 0.0634 44 H 2, 6 0.02, 0 0.01, 0 23.13 0.002 Synechococcus Abundance 20 H 2, 7 8.23 × 107, 4.11 × 107 4.11 × 107, 4.51 × 107 0.91 0.4509 44 H 2, 7 5.31 × 108, 6.97 × 107 2.65 × 108, 1.16 × 107 22.84 0.0016 Prochlorococcus Abundance 20 H 2, 8 4.22 × 107, 2.11 × 107 2.11 × 107, 2.71 × 106 7.77 0.0216 44 H 2, 8 9.02 × 107, 1.47 × 107 4.51 × 107, 2.45 × 106 18.38 0.0028 Pico-eukaryote -

Development of the Annelid Axochord: Insights Into Notochord Evolution Antonella Lauri Et Al
RESEARCH | REPORTS ORIGIN OF NOTOCHORD by double WMISH (Fig. 2, F to L). Although none of the genes were exclusively expressed in the annelid mesodermal midline, their combined Development of the annelid coexpression was unique to these cells (implying that mesodermal midline in annelids and chor- damesoderm in vertebrates are more similar to axochord: Insights into each other than to any other tissue). It is unlikely that the molecular similarity between annelid notochord evolution and vertebrate mesodermal midline is due to in- dependent co-option of a conserved gene cas- Antonella Lauri,1*† Thibaut Brunet,1* Mette Handberg-Thorsager,1,2‡ sette, because this would require either that this Antje H.L. Fischer,1§ Oleg Simakov,1 Patrick R. H. Steinmetz,1‖ Raju Tomer,1,2¶ cassette was active elsewhere in the body (which Philipp J. Keller,2 Detlev Arendt1,3# is not the case) or that multiple identical inde- pendent events of co-option occurred (which is The origin of chordates has been debated for more than a century, with one key issue being unparsimonious). As in vertebrates, the meso- the emergence of the notochord. In vertebrates, the notochord develops by convergence dermal midline resembles the neuroectodermal and extension of the chordamesoderm, a population of midline cells of unique molecular midline, which expresses foxD, foxA, netrin, slit, identity. We identify a population of mesodermal cells in a developing invertebrate, the marine and noggin (figs. S6 and S7) but not brachyury or annelid Platynereis dumerilii, that converges and extends toward the midline and expresses a twist. However, unlike in chicken (10), the an- notochord-specific combination of genes. -

Defining Phyla: Evolutionary Pathways to Metazoan Body Plans
EVOLUTION & DEVELOPMENT 3:6, 432-442 (2001) Defining phyla: evolutionary pathways to metazoan body plans Allen G. Collins^ and James W. Valentine* Museum of Paleontology and Department of Integrative Biology, University of California, Berkeley, CA 94720, USA 'Author for correspondence (email: [email protected]) 'Present address: Section of Ecology, Befiavior, and Evolution, Division of Biology, University of California, San Diego, La Jolla, CA 92093-0116, USA SUMMARY Phyla are defined by two sets of criteria, one pothesis of Nielsen; the clonal hypothesis of Dewel; the set- morphological and the other historical. Molecular evidence aside cell hypothesis of Davidson et al.; and a benthic hy- permits the grouping of animals into clades and suggests that pothesis suggested by the fossil record. It is concluded that a some groups widely recognized as phyla are paraphyletic, benthic radiation of animals could have supplied the ances- while some may be polyphyletic; the phyletic status of crown tral lineages of all but a few phyla, is consistent with molecu- phyla is tabulated. Four recent evolutionary scenarios for the lar evidence, accords well with fossil evidence, and accounts origins of metazoan phyla and of supraphyletic clades are as- for some of the difficulties in phylogenetic analyses of phyla sessed in the light of a molecular phylogeny: the trochaea hy- based on morphological criteria. INTRODUCTION Molecules have provided an important operational ad- vance to addressing questions about the origins of animal Concepts of animal phyla have changed importantly from phyla. Molecular developmental and comparative genomic their origins in the six Linnaean classis and four Cuvieran evidence offer insights into the genetic bases of body plan embranchements. -

Xenacoelomorpha's Significance for Understanding Bilaterian Evolution
Available online at www.sciencedirect.com ScienceDirect Xenacoelomorpha’s significance for understanding bilaterian evolution Andreas Hejnol and Kevin Pang The Xenacoelomorpha, with its phylogenetic position as sister biology models are the fruitfly Drosophila melanogaster and group of the Nephrozoa (Protostomia + Deuterostomia), plays the nematode Caenorhabditis elegans, in which basic prin- a key-role in understanding the evolution of bilaterian cell types ciples of developmental processes have been studied in and organ systems. Current studies of the morphological and great detail. It might be because the field of evolutionary developmental diversity of this group allow us to trace the developmental biology — EvoDevo — has its origin in evolution of different organ systems within the group and to developmental biology and not evolutionary biology that reconstruct characters of the most recent common ancestor of species under investigation are often called ‘model spe- Xenacoelomorpha. The disparity of the clade shows that there cies’. Criteria for selected representative species are cannot be a single xenacoelomorph ‘model’ species and primarily the ease of access to collected material and strategic sampling is essential for understanding the evolution their ability to be cultivated in the lab [1]. In some cases, of major traits. With this strategy, fundamental insights into the a supposedly larger number of ancestral characters or a evolution of molecular mechanisms and their role in shaping dominant role in ecosystems have played an additional animal organ systems can be expected in the near future. role in selecting model species. These arguments were Address used to attract sufficient funding for genome sequencing Sars International Centre for Marine Molecular Biology, University of and developmental studies that are cost-intensive inves- Bergen, Thormøhlensgate 55, 5008 Bergen, Norway tigations. -

Radial Symmetry Or Bilateral Symmetry Or "Spherical Symmetry"
Symmetry in biology is the balanced distribution of duplicate body parts or shapes. The body plans of most multicellular organisms exhibit some form of symmetry, either radial symmetry or bilateral symmetry or "spherical symmetry". A small minority exhibit no symmetry (are asymmetric). In nature and biology, symmetry is approximate. For example, plant leaves, while considered symmetric, will rarely match up exactly when folded in half. Radial symmetry These organisms resemble a pie where several cutting planes produce roughly identical pieces. An organism with radial symmetry exhibits no left or right sides. They have a top and a bottom (dorsal and ventral surface) only. Animals Symmetry is important in the taxonomy of animals; animals with bilateral symmetry are classified in the taxon Bilateria, which is generally accepted to be a clade of the kingdom Animalia. Bilateral symmetry means capable of being split into two equal parts so that one part is a mirror image of the other. The line of symmetry lies dorso-ventrally and anterior-posteriorly. Most radially symmetric animals are symmetrical about an axis extending from the center of the oral surface, which contains the mouth, to the center of the opposite, or aboral, end. This type of symmetry is especially suitable for sessile animals such as the sea anemone, floating animals such as jellyfish, and slow moving organisms such as sea stars (see special forms of radial symmetry). Animals in the phyla cnidaria and echinodermata exhibit radial symmetry (although many sea anemones and some corals exhibit bilateral symmetry defined by a single structure, the siphonoglyph) (see Willmer, 1990).