Fern Gazette V17 P4 V9.Qxd 01/02/2006 09:51 Page 235
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Guidance Document Pohakuloa Training Area Plant Guide
GUIDANCE DOCUMENT Recovery of Native Plant Communities and Ecological Processes Following Removal of Non-native, Invasive Ungulates from Pacific Island Forests Pohakuloa Training Area Plant Guide SERDP Project RC-2433 JULY 2018 Creighton Litton Rebecca Cole University of Hawaii at Manoa Distribution Statement A Page Intentionally Left Blank This report was prepared under contract to the Department of Defense Strategic Environmental Research and Development Program (SERDP). The publication of this report does not indicate endorsement by the Department of Defense, nor should the contents be construed as reflecting the official policy or position of the Department of Defense. Reference herein to any specific commercial product, process, or service by trade name, trademark, manufacturer, or otherwise, does not necessarily constitute or imply its endorsement, recommendation, or favoring by the Department of Defense. Page Intentionally Left Blank 47 Page Intentionally Left Blank 1. Ferns & Fern Allies Order: Polypodiales Family: Aspleniaceae (Spleenworts) Asplenium peruvianum var. insulare – fragile fern (Endangered) Delicate ENDEMIC plants usually growing in cracks or caves; largest pinnae usually <6mm long, tips blunt, uniform in shape, shallowly lobed, 2-5 lobes on acroscopic side. Fewer than 5 sori per pinna. Fronds with distal stipes, proximal rachises ocassionally proliferous . d b a Asplenium trichomanes subsp. densum – ‘oāli’i; maidenhair spleenwort Plants small, commonly growing in full sunlight. Rhizomes short, erect, retaining many dark brown, shiny old stipe bases.. Stipes wiry, dark brown – black, up to 10cm, shiny, glabrous, adaxial surface flat, with 2 greenish ridges on either side. Pinnae 15-45 pairs, almost sessile, alternate, ovate to round, basal pinnae smaller and more widely spaced. -
Asplenium Bulbiferum
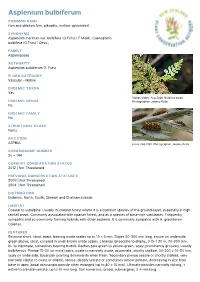
Asplenium bulbiferum COMMON NAME Hen and chicken fern, pikopiko, mother spleenwort SYNONYMS Asplenium marinum var. bulbifera (G.Forst.) F.Muell.; Caenopteris bulbifera (G.Forst.) Desv.; FAMILY Aspleniaceae AUTHORITY Asplenium bulbiferum G. Forst. FLORA CATEGORY Vascular – Native ENDEMIC TAXON Yes Stokes Valley. Aug 2006. Bulbil on bulbil. ENDEMIC GENUS Photographer: Jeremy Rolfe No ENDEMIC FAMILY No STRUCTURAL CLASS Ferns NVS CODE ASPBUL sorus. Feb 1981. Photographer: Jeremy Rolfe CHROMOSOME NUMBER 2n = 144 CURRENT CONSERVATION STATUS 2012 | Not Threatened PREVIOUS CONSERVATION STATUSES 2009 | Not Threatened 2004 | Not Threatened DISTRIBUTION Endemic. North, South, Stewart and Chatham Islands HABITAT Coastal to subalpine. Usually in lowland forest where it is a common species of the ground-layer, especially in high rainfall areas. Commonly associated with riparian forest, and as a species of base-rich substrates. Frequently sympatric and so commonly forming hybrids with other asplenia. It is commonly sympatric with A. gracillimum Colenso. FEATURES Rhizome short, stout, erect, bearing ovate scales up to 15 × 5 mm. Stipes 50-300 mm long, brown on underside, green above, stout, covered in small brown ovate scales. Laminae lanceolate to elliptic, 0.15-1.20 m, 70-300 mm, bi- to tripinnate, sometimes bearing bulbils. Raches pale green to yellow-green, scaly, prominently grooved, usually bulbiferous. Pinnae 15-30 (or more) pairs, ovate to narrowly ovate, acuminate, shortly stalked, 30-200 × 10-50 mm, scaly on underside, basal pair pointing downwards when fresh. Secondary pinnae sessile or shortly stalked, very narrowly elliptic to ovate or elliptic, obtuse, deeply serrate or sometimes almost pinnate, decreasing in size from base to apex, basal acroscopic pinnule often enlarged (up to 40 × 10 mm). -

Ferns Robert H
Southern Illinois University Carbondale OpenSIUC Illustrated Flora of Illinois Southern Illinois University Press 10-1999 Ferns Robert H. Mohlenbrock Southern Illinois University Carbondale Follow this and additional works at: http://opensiuc.lib.siu.edu/siupress_flora_of_illinois Part of the Botany Commons Recommended Citation Mohlenbrock, Robert H., "Ferns" (1999). Illustrated Flora of Illinois. 3. http://opensiuc.lib.siu.edu/siupress_flora_of_illinois/3 This Book is brought to you for free and open access by the Southern Illinois University Press at OpenSIUC. It has been accepted for inclusion in Illustrated Flora of Illinois by an authorized administrator of OpenSIUC. For more information, please contact [email protected]. THE ILLUSTRATED FLORA OF ILLINOIS ROBERT H. MOHLENBROCK, General Editor THE ILLUSTRATED FLORA OF ILLINOIS s Second Edition Robert H. Mohlenbrock SOUTHERN ILLINOIS UNIVERSITY PRESS Carbondale and Edwardsville COPYRIGHT© 1967 by Southern Illinois University Press SECOND EDITION COPYRIGHT © 1999 by the Board of Trustees, Southern Illinois University All rights reserved Printed in the United States of America 02 01 00 99 4 3 2 1 Library of Congress Cataloging-in-Publication Data Mohlenbrock, Robert H., 1931- Ferns I Robert H. Mohlenbrock. - 2nd ed. p. em.- (The illustrated flora of Illinois) Includes bibliographical references and index. 1. Ferns-Illinois-Identification. 2. Ferns-Illinois-Pictorial works. 3. Ferns-Illinois-Geographical distribution-Maps. 4. Botanical illustration. I. Title. II. Series. QK525.5.I4M6 1999 587'.3'09773-dc21 99-17308 ISBN 0-8093-2255-2 (cloth: alk. paper) CIP The paper used in this publication meets the minimum requirements of American National Standard for Information Sciences-Permanence of Paper for Printed Library Materials, ANSI Z39.48-1984.§ This book is dedicated to Miss E. -

Mississippi Natural Heritage Program Special Plants - Tracking List -2018
MISSISSIPPI NATURAL HERITAGE PROGRAM SPECIAL PLANTS - TRACKING LIST -2018- Approximately 3300 species of vascular plants (fern, gymnosperms, and angiosperms), and numerous non-vascular plants may be found in Mississippi. Many of these are quite common. Some, however, are known or suspected to occur in low numbers; these are designated as species of special concern, and are listed below. There are 495 special concern plants, which include 4 non- vascular plants, 28 ferns and fern allies, 4 gymnosperms, and 459 angiosperms 244 dicots and 215 monocots. An additional 100 species are designated “watch” status (see “Special Plants - Watch List”) with the potential of becoming species of special concern and include 2 fern and fern allies, 54 dicots and 44 monocots. This list is designated for the primary purposes of : 1) in environmental assessments, “flagging” of sensitive species that may be negatively affected by proposed actions; 2) determination of protection priorities of natural areas that contain such species; and 3) determination of priorities of inventory and protection for these plants, including the proposed listing of species for federal protection. GLOBAL STATE FEDERAL SPECIES NAME COMMON NAME RANK RANK STATUS BRYOPSIDA Callicladium haldanianum Callicladium Moss G5 SNR Leptobryum pyriforme Leptobryum Moss G5 SNR Rhodobryum roseum Rose Moss G5 S1? Trachyxiphium heteroicum Trachyxiphium Moss G2? S1? EQUISETOPSIDA Equisetum arvense Field Horsetail G5 S1S2 FILICOPSIDA Adiantum capillus-veneris Southern Maidenhair-fern G5 S2 Asplenium -

Oakes and Camptosorus Rhizophyllus
A STUDY OF As?lenium platyneuron (L.) Oakes AND Camptosorus rhizophyllus (L.) Link \-l1TH AN .SllPH..t::..2IS em SPOiC:': 10Rl.~HOLOGY A Senior Paper Submitted to Or. J. C. r.falayer of Ball State University by Lois A. I(inder In Partial Fulfillment of the Requiren.ents for graduation on The Honors Program l'.J:ay I, 1966 :;;rCo~1 7he:! ii ":'"\ J-i-t.:., (.~1 ~ ' ..... -, ~--~ ~: i,~ , '")6 t, ,k ,~-r;0 TABLE OF CONTENTS Page LIST OF TABLES ••••••••••.••••.••••••••••••••.••..• iii LIST OF ILLUSTRATIONS. .. .. .. .. iv INTRODUCTION •••••••••••••••• .. .. .. 1 REVIE\~ OF irH~~ LI Ir .2.PJ.l.TUl<'E •••••••••••••••••••••••••• 2 Morphology ••••.•••••..•••••••••••••••••••••••••• 2 Taxonomic Realtionships •• ••••••••••••••••••••••• 4 NETrIODS MATERlii.LS •• .. .. .. .. .. 6 General 1>1orphology •••••••••••••••••••••••••••••• 6 :-")pore }forphology •••••••••••••••••••••••••••••••• 7 '[lATA ••••••••••••••••• . .. 15 General Horphology.............................. 15 Spore Morphology................................ 30 DISCUSSION. .. .. .. .. .. .. .. .. 39 sm~J~y........................................... 46 BIBLIOGRAPHY...................................... 47 iii LIST OF TABLES Page Table I. Gross morphological calculations of Gamptosorus rhizophyllus (L.) Link I. A Specimen measurements ••••••••••••••••• 8 I. B Leaf height analysis •••••••••••••••••• 19 II. Gross morphological calculations of iisplenium platyneuron (L.) C.akes II. A Specimen measurements................. 9 II. B Leaf height analysis ••••••••••••••••• 24 III. :}ross morphological -

Asplenium Rhizophyllum L
Asplenium rhizophyllum L. walking fern Photos by Michael R. Penskar State Distribution Best Survey Period Jan Feb Mar Apr May Jun Jul Aug Sep Oct Nov Dec Status: State threatened Niagara Escarpment. Elsewhere, this species occurs locally on alkaline bedrock outcrops in Dickinson, Global and state rank: G5/S2S3 Schoolcraft, and Houghton counties, with additional Family: Aspleniaceae (spleenwort family) local populations found in the Lower Peninsula on South Manitou Island (Leleenau County). It is also Synonyms: Camptosorus rhizophyllus (L.) Link known from a sinkhole in Alpena County, and from an unusual occurrence in Berrien County where a small Taxonomy: This very distinctive species has been but vigorous colony was discovered on a limestone segregated by several authors and placed in the genus boulder along a stream. Camptosorus (Morin et al. 1993), a name under which it is known in many manuals and other publications. It Recognition: Asplenium rhizophyllum is an extremely forms part of a complex of Appalachian spleenworts distinctive fern, characterized by its tendency to form researched by Wagner (1954) in a well-known study of dense colonies by reproducing via tip-rooting on hybridization and backcrossing. moss-covered dolomite boulders and other types of rock outcrops. Individual plants consist of clumps of Total range: Walking fern occurs in eastern North fronds (leaves) arising from short, scaly rhizomes. The America, ranging from southern Ontario and Quebec small, 1-3 cm wide fronds, which have net-like (reticu- in Canada south to Georgia, Alabama, and Mississippi, late) veins and may range up to ca. 30 cm in length, occurring west to Wisconsin, Iowa, Kansas, and are stalked, and have slender, long-tapering, lance- Oklahoma. -

100 Years of Change in the Flora of the Carolinas
Flora of the Carolinas, Virginia, Georgia, and surrounding areas Working Draft of 11 January 2007 by Alan S. Weakley University of North Carolina Herbarium (NCU) North Carolina Botanical Garden University of North Carolina at Chapel Hill Campus Box 3280 Chapel Hill NC 27599-3280 TABLE OF CONTENTS Table of Contents THE FLORA .................................................................................................................................................................................................................6 ACKNOWLEDGMENTS ............................................................................................................................................................................................9 FERNS AND “FERN ALLIES”............................................................................................................................................................................10 ASPLENIACEAE Frank 1877 (Spleenwort Family) ........................................................................................................................................17 AZOLLACEAE Wettstein 1903 (Mosquito Fern Family) .................................................................................................................................20 BLECHNACEAE (C. Presl) Copeland 1947 (Deer Fern Family) ...................................................................................................................21 DENNSTAEDTIACEAE Pichi Sermolli 1970 (Bracken Family) ....................................................................................................................22 -

PACIFIC INSECTS MONOGRAPH Ll
PACIFIC INSECTS MONOGRAPH ll Lepidoptera of American Samoa with particular reference to biology and ecology By John Adams Comstock Published by Entomology Department, Bernice P. Bishop Museum Honolulu, Hawaii, U. S. A. 1966 PACIFIC INSECTS MONOGRAPHS Published by Entomology Department, Bernice P. Bishop Museum, Honolulu, Hawaii, 96819, U. S. A. Editorial Committee: J. L. Gressitt, Editor (Honolulu), S. Asahina (Tokyo), R. G. Fennah (London), R. A. Harrison (Christchurch), T. C. Maa (Honolulu & Taipei), C. W. Sabrosky (Washington, D. C), R. L. Usinger (Berkeley), J. van der Vecht (Leiden), K. Yasumatsu (Fukuoka), E. C. Zimmerman (New Hampshire). Assistant Editors: P. D. Ashlock (Honolulu), Carol Higa (Honolulu), Naoko Kunimori (Fukuoka), Setsuko Nakata (Honolulu), Toshi Takata (Fukuoka). Business Manager: C. M. Yoshimoto (Honolulu). Business Assistant: Doris Anbe (Honolulu). Business Agent in Japan: K. Yasumatsu (Fukuoka). Entomological staff, Bishop Museum, 1966: Doris Anbe, Hatsuko Arakaki, P. D. Ashlock, S. Azuma, Madaline Boyes, Candida Cardenas, Ann Cutting, M. L. Goff, J. L. Gressitt (Chairman), J. Harrell, Carol Higa, Y. Hirashima, Shirley Hokama, E. Holzapfel, Dorothy Hoxie, Helen Hurd, June Ibara, Naoko Kuni mori, T. C. Maa, Grace Nakahashi, Setsuko Nakata (Adm. Asst.), Tulene Nonomura, Carol Okuma, Ka tharine Pigue, Linda Reineccius, T. Saigusa, I. Sakakibara, Judy Sakamoto, G. A. Samuelson, Sybil Seto, W. A. Steffan, Amy Suehiro, Grace Thompson, Clara Uchida, J. R. Vockeroth, Nixon Wilson, Mabel Ya- tsuoka, C. M. Yoshimoto, E. C. Zimmermann. Field associates: M. J. Fitzsimons, E. E. Gless, G. E. Lip- pert, V. Peckham, D. S. Rabor, J. Sedlacek, M. Sedlacek, P. Shanahan, R. Straatman, J. Strong, H. M. Tor- revillas, A. -

New and Noteworthy Additions to the Arkansas Fern Flora
Peck, J.H. 2011. New and noteworthy additions to the Arkansas fern flora. Phytoneuron 2011-30: 1–33. NEW AND NOTEWORTHY ADDITIONS TO THE ARKANSAS FERN FLORA JAMES H. PECK Department Biology University of Arkansas at Little Rock 2801 S. University Ave. Little Rock, AR 72204 [email protected] ABSTRACT Since 1995, 11 fern taxa have been added to the Arkansas flora as new and native, including Asplenium montanum , A. ruta-muraria , A. septentrianale , A. ×trudellii , Athyrium angustum , Azolla caroliniana , Dryopteris goldiana , D. celsa × goldiana , Marsilea macropoda , Palhinhaea cernua , and Trichomanes intracatum . Of the reported Arkansas native ferns, one was deleted (Azolla caroliniana ), being subsumed by Azolla mexicana and now correctly known as Azolla microphylla . Since 1995, 20 fern taxa have been added to the Arkansas fern flora as new and naturalized, including Arachnioides simplicior , Athyrium nipponicum ‘Pictum’, Cyrtomium falcatum , C. fortunei , Dryopteris erythrospora , Hypolepis tenuifolia , Marsilea mutica , M. quadrifolia , Matteuccia struthiopteris , Nephrolepis exaltata , Polystichum tsus-sinense , Phegopteris decursive-pinnata , Salvinia minima , S. molesta , Selaginella braunii , S. kraussiana , S. k. ‘Aurea’, S. k. ‘Brownii’, S. k. ‘Goldtips’, and S. uncinata . Of the reported Arkansas naturalized ferns, one was deleted (C. fortunei ), being without a known voucher. There are now 97 native and 24 naturalized fern taxa known and documented in the Arkansas fern flora. The total Arkansas fern flora is now 121 taxa documented with 3019 county-level occurrence records. Noteworthy update records and comments are reported for 79 of 97 Arkansas native species and 25 Arkansas naturalized species. KEY WORDS : Arkansas, ferns, county distribution Over the last 30 years, studies have been conducted to document the diversity and abundance of the Arkansas fern [pteridophyte] flora. -

HARDY FERN FOUNDATION QUARTERLY the HARDY FERN FOUNDATION Quarterly Volume 8 • No
THE HARDY FERN FOUNDATION P.O. Box 166 Medina, WA 98039-0166 [email protected] Web site darkwing, uoregon .edu/~sueman/ The Hardy Fern Foundation was founded in 1989 to establish a comprehensive collection of the world’s hardy ferns for display, testing, evaluation, public educa¬ tion and introduction to the gardening and horticultural community. Many rare and unusual species, hybrids and varieties are being propagated from spores and tested in selected environments for their different degrees of hardiness and orna¬ mental garden value. The primary fern display and test garden is located at, and in conjunction with, The Rhododendron Species Botanical Garden at the Weyerhaeuser Corporate Headquarters, in Federal Way, Washington. Satellite fern gardens are at the Stephen Austin Arboretum, Nacogdoches, Texas, Birmingham Botanical Gardens, Birmingham, Alabama, California State Univer¬ sity at Sacramento, Sacramento, California, Dallas Arboretum, Dallas, Texas, Denver Botanic Gardens. Denver, Colorado, Georgeson Botanical Garden, Uni¬ versity of Alaska, Fairbanks, Alaska, Harry P. Leu Garden, Orlando, Florida, Coastal Maine Botanical Garden, Wiscasset, Maine, Inniswood Metro Gardens, Colum¬ bus, Ohio, New York Botanical Garden, Bronx, New York, and Strybing Arbore¬ tum, San Francisco, California. The fern display gardens are at Lakewold, Tacoma, Washington, Les Jardins de Metis, Quebec, Canada, University of Northern Colorado, Greeley, Colorado, and Whitehall Historic Home and Garden, Louisville, KY. Hardy Fern Foundation members participate in a spore exchange, receive a quar¬ terly newsletter and have first access to ferns as they are ready for distribution. Cover Design by Willanna Bradner. HARDY FERN FOUNDATION QUARTERLY THE HARDY FERN FOUNDATION Quarterly Volume 8 • No. 3 • Editor Sue Olsen \ T *2 W4 g WS11 U President’s Message.47 Anne C. -

Conservation Assessment for Walking Fern (Asplenium Rhizophyllum) L
Conservation Assessment for Walking fern (Asplenium rhizophyllum) L. Photo: Arien Tal USDA Forest Service, Region 9 March 2002 This document is undergoing peer review, comments welcome This Conservation Assessment was prepared to compile the published and unpublished information on the subject taxon or community; or this document was prepared by another organization and provides information to serve as a Conservation Assessment for the Eastern Region of the Forest Service. It does not represent a management decision by the U.S. Forest Service. Though the best scientific information available was used and subject experts were consulted in preparation of this document, it is expected that new information will arise. In the spirit of continuous learning and adaptive management, if you have information that will assist in conserving the subject taxon, please contact the Eastern Region of the Forest Service - Threatened and Endangered Species Program at 310 Wisconsin Avenue, Suite 580 Milwaukee, Wisconsin 53203. Conservation Assessment forWalking fern (Asplenium rhizophyllum L.) 2 Table of Contents ACKNOWLEDGEMENTS ......................................................................... 4 EXECUTIVE SUMMARY ......................................................................... 4 NOMENCLATURE AND TAXONOMY (W-1)........................................ 5 DESCRIPTION OF SPECIES .................................................................... 5 HABITAT AND ECOLOGY....................................................................... 6 DISTRIBUTION -

Supplementary Table 1
Supplementary Table 1 SAMPLE CLADE ORDER FAMILY SPECIES TISSUE TYPE CAPN Eusporangiate Monilophytes Equisetales Equisetaceae Equisetum diffusum developing shoots JVSZ Eusporangiate Monilophytes Equisetales Equisetaceae Equisetum hyemale sterile leaves/branches NHCM Eusporangiate Monilophytes Marattiales Marattiaceae Angiopteris evecta developing shoots UXCS Eusporangiate Monilophytes Marattiales Marattiaceae Marattia sp. leaf BEGM Eusporangiate Monilophytes Ophioglossales Ophioglossaceae Botrypus virginianus Young sterile leaf tissue WTJG Eusporangiate Monilophytes Ophioglossales Ophioglossaceae Ophioglossum petiolatum leaves, stalk, sporangia QHVS Eusporangiate Monilophytes Ophioglossales Ophioglossaceae Ophioglossum vulgatum EEAQ Eusporangiate Monilophytes Ophioglossales Ophioglossaceae Sceptridium dissectum sterile leaf QVMR Eusporangiate Monilophytes Psilotales Psilotaceae Psilotum nudum developing shoots ALVQ Eusporangiate Monilophytes Psilotales Psilotaceae Tmesipteris parva Young fronds PNZO Cyatheales Culcitaceae Culcita macrocarpa young leaves GANB Cyatheales Cyatheaceae Cyathea (Alsophila) spinulosa leaves EWXK Cyatheales Thyrsopteridaceae Thyrsopteris elegans young leaves XDVM Gleicheniales Gleicheniaceae Sticherus lobatus young fronds MEKP Gleicheniales Dipteridaceae Dipteris conjugata young leaves TWFZ Hymenophyllales Hymenophyllaceae Crepidomanes venosum young fronds QIAD Hymenophyllales Hymenophyllaceae Hymenophyllum bivalve young fronds TRPJ Hymenophyllales Hymenophyllaceae Hymenophyllum cupressiforme young fronds and sori