Microrna Deregulation in Papillary Thyroid Cancer and Its
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Mechanism of Activation of Monomeric B-Raf V600E
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.06.438646; this version posted June 8, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The Mechanism of Activation of Monomeric B-Raf V600E Ryan C. Maloneya, Mingzhen Zhanga, Hyunbum Janga and Ruth Nussinova,b,* a Computational Structural Biology Section, Frederick National Laboratory for Cancer Research in the Laboratory of Cancer Immunometabolism, National Cancer Institute, Frederick, MD 21702, U.S.A b Department of Human Molecular Genetics and Biochemistry, Sackler School of Medicine, Tel Aviv University, Tel Aviv 69978, Israel * Author for correspondence: Tel: 1-301-846-5579 E-mail: [email protected] Declarations of interest: none 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.04.06.438646; this version posted June 8, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract Oncogenic mutations in the serine/threonine kinase B-Raf, particularly the V600E mutation, are frequent in cancer, making it a major drug target. Although much is known about B-Raf’s active and inactive states, questions remain about the mechanism by which the protein changes between these two states. Here, we utilize molecular dynamics to investigate both wild-type and V600E B-Raf to gain mechanistic insights into the impact of the Val to Glu mutation. -

1596.Full-Text.Pdf
Published OnlineFirst May 12, 2017; DOI: 10.1158/1535-7163.MCT-16-0798 Cancer Biology and Signal Transduction Molecular Cancer Therapeutics Identification of the Serine Biosynthesis Pathway as a Critical Component of BRAF Inhibitor Resistance of Melanoma, Pancreatic, and Non–Small Cell Lung Cancer Cells Kayleigh C. Ross1, Andrew J. Andrews1,2, Christopher D. Marion1, Timothy J. Yen2, and Vikram Bhattacharjee1 Abstract Metastatic melanoma cells commonly acquire resistance to to BRAF WT tumor cell lines that are intrinsically resistant to BRAF V600E inhibitors (BRAFi). In this study, we identified vemurafenib and dabrafenib. Pretreatment of pancreatic serine biosynthesis as a critical mechanism of resistance. Prote- cancer and non–small cell lung cancer cell lines with sublethal omic assays revealed differential protein expression of serine dosesof50and5nmol/Lofgemcitabine,respectively, biosynthetic enzymes PHGDH, PSPH, and PSAT1 following enhanced killing by both vemurafenib and dabrafenib. The vemurafenib (BRAFi) treatment in sensitive versus acquired novel aspects of this study are the direct identification of serine resistant melanoma cells. Ablation of PHGDH via siRNA sen- biosynthesis as a critical mechanism of BRAF V600E inhibitor sitizedacquiredresistantcells to vemurafenib. Inhibiting the resistance and the first successful example of using gemcitabine folate cycle, directly downstream of serine synthesis, with þ BRAFis in combination to kill previously drug-resistant methotrexate also displayed similar sensitization. Using cancer cells, creating the translational potential of pretreatment the DNA-damaging drug gemcitabine, we show that gemcita- with gemcitabine prior to BRAFi treatment of tumor cells to bine pretreatment sensitized resistant melanoma cells to BRA- reverse resistance within the mutational profile and the WT. Fis vemurafenib and dabrafenib. -

COTELLIC™ (Cobimetinib) Oral Tablet
PHARMACY COVERAGE GUIDELINES ORIGINAL EFFECTIVE DATE: 1/21/2016 SECTION: DRUGS LAST REVIEW DATE: 2/18/2021 LAST CRITERIA REVISION DATE: 2/18/2021 ARCHIVE DATE: COTELLIC™ (cobimetinib) oral tablet Coverage for services, procedures, medical devices and drugs are dependent upon benefit eligibility as outlined in the member's specific benefit plan. This Pharmacy Coverage Guideline must be read in its entirety to determine coverage eligibility, if any. This Pharmacy Coverage Guideline provides information related to coverage determinations only and does not imply that a service or treatment is clinically appropriate or inappropriate. The provider and the member are responsible for all decisions regarding the appropriateness of care. Providers should provide BCBSAZ complete medical rationale when requesting any exceptions to these guidelines. The section identified as “Description” defines or describes a service, procedure, medical device or drug and is in no way intended as a statement of medical necessity and/or coverage. The section identified as “Criteria” defines criteria to determine whether a service, procedure, medical device or drug is considered medically necessary or experimental or investigational. State or federal mandates, e.g., FEP program, may dictate that any drug, device or biological product approved by the U.S. Food and Drug Administration (FDA) may not be considered experimental or investigational and thus the drug, device or biological product may be assessed only on the basis of medical necessity. Pharmacy Coverage Guidelines are subject to change as new information becomes available. For purposes of this Pharmacy Coverage Guideline, the terms "experimental" and "investigational" are considered to be interchangeable. BLUE CROSS®, BLUE SHIELD® and the Cross and Shield Symbols are registered service marks of the Blue Cross and Blue Shield Association, an association of independent Blue Cross and Blue Shield Plans. -

Distinguishing Clinicopathologic Features of Patients with V600E and V600K BRAF-Mutant Metastatic Melanoma
Published OnlineFirst April 24, 2012; DOI: 10.1158/1078-0432.CCR-12-0052 Clinical Cancer Human Cancer Biology Research Distinguishing Clinicopathologic Features of Patients with V600E and V600K BRAF-Mutant Metastatic Melanoma Alexander M. Menzies1,2,6, Lauren E. Haydu1,3, Lydia Visintin1,6,7, Matteo S. Carlino1,2,5,6, Julie R. Howle1,3,6, John F. Thompson1,3,7,8, Richard F. Kefford1,2,5,6, Richard A. Scolyer1,4,7, and Georgina V. Long1,2,6 Abstract Purpose: Certain clinicopathologic features correlate with BRAF mutation status in melanoma including younger age and primary subtype. This study sought to determine the BRAF mutation status by age-decade and whether BRAF-mutant genotypes correlated with clinicopathologic features and outcome in patients with metastatic melanoma. Methods: A prospectively assembled cohort of Australian patients were followed from diagnosis of metastatic melanoma (N ¼ 308). Clinicopathologic variables were correlated with BRAF mutational status, genotype, and survival. Results: Forty-six percent of patients had a BRAF mutation; 73% V600E, 19% V600K, and 8% other genotypes. An inverse relationship existed between BRAF mutation prevalence and age-decade (P < 0.001). All patients <30 years and only 25% 70 years had BRAF-mutant melanoma. Amongst BRAF-mutant melanoma, the frequency of non-V600E genotypes (including V600K) increased with increasing age. Non- V600E genotypes comprised <20% in patients <50 years and >40% in those 70 years. A higher degree of cumulative sun-induced damage correlated with V600K but not V600E melanoma (P ¼ 0.002). The disease- free interval from diagnosis of primary melanoma to first distant metastasis was shorter for patients with V600K compared with V600E melanoma (17.4 vs. -

Genetic Landscape of Adult Langerhans Cell Histiocytosis with Lung Involvement
Early View Original article Genetic Landscape of Adult Langerhans Cell Histiocytosis with Lung Involvement Fanélie Jouenne, Sylvie Chevret, Emmanuelle Bugnet, Emmanuelle Clappier, Gwenaël Lorillon, Véronique Meignin, Aurélie Sadoux, Shannon Cohen, Alain Haziot, Alexandre How-Kit, Caroline Kannengiesser, Céleste Lebbé, Dominique Gossot, Samia Mourah, Abdellatif Tazi Please cite this article as: Jouenne Félie, Chevret S, Bugnet E, et al. Genetic Landscape of Adult Langerhans Cell Histiocytosis with Lung Involvement. Eur Respir J 2019; in press (https://doi.org/10.1183/13993003.01190-2019). This manuscript has recently been accepted for publication in the European Respiratory Journal. It is published here in its accepted form prior to copyediting and typesetting by our production team. After these production processes are complete and the authors have approved the resulting proofs, the article will move to the latest issue of the ERJ online. Copyright ©ERS 2019 Genetic Landscape of Adult Langerhans Cell Histiocytosis with Lung Involvement Fanélie Jouenne,1,2 Sylvie Chevret,3,4 Emmanuelle Bugnet,5 Emmanuelle Clappier,6,7 Gwenaël Lorillon,5 Véronique Meignin,8 Aurélie Sadoux,2 Shannon Cohen,9 Alain Haziot,9 Alexandre How-Kit,10 Caroline Kannengiesser,11 Céleste Lebbé,1,12 Dominique Gossot,13 Samia Mourah,1,2 and Abdellatif Tazi1,5,* 1Université de Paris, INSERM U976, Institut de Recherche Saint-Louis, Paris, France 2Assistance Publique-Hôpitaux de Paris, Hôpital Saint-Louis, Laboratoire de Pharmacogénomique, Paris, France 3Université de Paris, -

COTELLIC (Cobimetinib) RATIONALE for INCLUSION in PA PROGRAM
COTELLIC (cobimetinib) RATIONALE FOR INCLUSION IN PA PROGRAM Background Cotellic (cobimetinib) is indicated for the treatment of patients with metastatic or unresectable melanoma, a type of skin cancer. Cotellic is specifically indicated for the treatment of patients with melanoma whose tumors express a gene mutation called BRAF V600E or V600K. The mutated BRAF protein signals the melanoma cells to replicate faster. Cotellic is taken in combination with vemurafenib. Cotellic and vemurafenib target two different proteins in the cell replication pathway, resulting in cell death and reduction of tumor growth. Cotellic is administered, in combination with vemurafenib, once a day for the first 21 days of each 28-day cycle until disease progression or unacceptable toxicity (1). Regulatory Status FDA-approved indication: Cotellic is a kinase inhibitor indicated for the treatment of patients with unresectable or metastatic melanoma with a BRAF V600E or V600K mutation, in combination with vemurafenib (1). Patients should be monitored for new malignancies prior to initiation of therapy, while on therapy, and for up to 6 months following the last dose of Cotellic (1). Major hemorrhagic (bleeding) events can occur with Cotellic. Monitor for signs and symptoms of bleeding (1). The risk of cardiomyopathy is increased in patients receiving the combination of Cotellic with vemurafenib. The safety of Cotellic has not been established in patients with decreased left ventricular ejection fraction. Left ventricular ejection fraction (LVEF) should be evaluated before treatment, after one month of treatment then every 3 months thereafter during treatment with Cotellic (1). Prescriber should perform an ophthalmological evaluation at regular intervals and for any visual disturbances. -

Cotellic® (Cobimetinib) P&T Approval Date 1/2016, 12/2016, 11/2017, 11/2018, 11/2019, 11/2020 Effective Date 2/1/2021; Oxford Only: 2/1/2021
UnitedHealthcare Pharmacy Clinical Pharmacy Programs Program Number 2020 P 1175-6 Program Prior Authorization/Notification Medication Cotellic® (cobimetinib) P&T Approval Date 1/2016, 12/2016, 11/2017, 11/2018, 11/2019, 11/2020 Effective Date 2/1/2021; Oxford only: 2/1/2021 1. Background: Cotellic® (cobimetinib) is a kinase inhibitor indicated for the treatment of patients with unresectable or metastatic melanoma with a BRAF V600E or V600K mutation, in combination with Zelboraf® (vemurafenib).1 The National Cancer Comprehensive Network (NCCN) also recommends the use of Cotellic in combination with Zelboraf® (vemurafenib) as treatment for Central Nervous System (CNS) Cancers. Coverage Information: Members will be required to meet the criteria below for coverage. For members under the age of 19 years, the prescription will automatically process without a coverage review. Some states mandate benefit coverage for off-label use of medications for some diagnoses or under some circumstances. Some states also mandate usage of other Compendium references. Where such mandates apply, they supersede language in the benefit document or in the notification criteria. 2. Coverage Criteria: A. Patients less than 19 years of age 1. Cotellic will be approved based on the following criterion: a. Patient is less than 19 years of age Authorization will be issued for 12 months. B. Melanoma 1. Initial Authorization a. Cotellic will be approved based on all of the following criteria: (1) Diagnosis of melanoma © 2020 UnitedHealthcare Services, Inc. 1 -AND- (2) Disease is one of the following: (a) Unresectable (b) Metastatic -AND- (3) Disease is positive for one of the following mutations: (a) BRAF V600E (b) BRAF V600K -AND- (4) Used in combination with Zelboraf (vemurafenib) Authorization will be issued for 12 months. -

Dabrafenib and Trametinib Treatment in Patients with Locally Advanced Or Metastatic BRAF V600–Mutant Anaplastic Thyroid Cancer Vivek Subbiah, Robert J
VOLUME 36 • NUMBER 1 • JANUARY 1, 2018 JOURNAL OF CLINICAL ONCOLOGY RAPID COMMUNICATION Dabrafenib and Trametinib Treatment in Patients With Locally Advanced or Metastatic BRAF V600–Mutant Anaplastic Thyroid Cancer Vivek Subbiah, Robert J. Kreitman, Zev A. Wainberg, Jae Yong Cho, Jan H.M. Schellens, Jean Charles Soria, Patrick Y. Wen, Christoph Zielinski, Maria E. Cabanillas, Gladys Urbanowitz, Bijoyesh Mookerjee, Dazhe Wang, Fatima Rangwala, and Bhumsuk Keam Author affiliations and support information (if applicable) appear at the end of this ABSTRACT article. Purpose Published at jco.org on October 26, 2017. We report the efficacy and safety of dabrafenib (BRAF inhibitor) and trametinib (MEK inhibitor) Processed as a Rapid Communication combination therapy in BRAF V600E–mutated anaplastic thyroid cancer, a rare, aggressive, and manuscript. highly lethal malignancy with poor patient outcomes and no systemic therapies with clinical benefit. Clinical trial information: NCT02034110. Methods Corresponding author: Bhumsuk Keam, In this phase II, open-label trial, patients with predefined BRAF V600E–mutated malignancies re- MD, PhD, Department of Internal Medicine, Seoul National University ceived dabrafenib 150 mg twice daily and trametinib 2 mg once daily until unacceptable toxicity, Hospital, 101 Daehak-ro, Jongno-gu, disease progression, or death. The primary end point was investigator-assessed overall response Seoul, Republic of Korea 110-744; e-mail: rate. Secondary end points included duration of response, progression-free survival, overall survival, [email protected]. and safety. © 2017 by American Society of Clinical Results Oncology Sixteen patients with BRAF V600E–mutated anaplastic thyroid cancer were evaluable (median 0732-183X/18/3601w-7w/$20.00 follow-up, 47 weeks; range, 4 to 120 weeks). -

1272.Full.Pdf
Published OnlineFirst January 10, 2019; DOI: 10.1158/1078-0432.CCR-18-1680 Translational Cancer Mechanisms and Therapy Clinical Cancer Research Distinct Molecular Profiles and Immunotherapy Treatment Outcomes of V600E and V600K BRAF-Mutant Melanoma Ines^ Pires da Silva1, Kevin Y.X. Wang2, James S. Wilmott1, Jeff Holst3, Matteo S. Carlino1,4, John J. Park5, Camelia Quek1, Matthew Wongchenko6, Yibing Yan6, Graham Mann1,7, Douglas B. Johnson8, Jennifer L. McQuade9, Rajat Rai1, Richard F. Kefford1,4,5, Helen Rizos1,5, Richard A. Scolyer1,10, Jean Y.H.Yang2, Georgina V. Long1,11, and Alexander M. Menzies1,11 Abstract Purpose: BRAF V600E and V600K melanomas have distinct lower expression of the ERK pathway feedback regulator clinicopathologic features, and V600K appear to be less dual-specificity phosphatase 6, confirmed with TCGA responsive to BRAFiÆMEKi. We investigated mechanisms for data (116 V600E, 17 V600K). Pathway analysis showed this and explored whether genotype affects response to V600K had lower expression of ERK and higher expression immunotherapy. of PI3K-AKT genes than V600E. Higher mutational load Experimental Design: Pretreatment formalin-fixed was observed in V600K, with a higher proportion of muta- paraffin-embedded tumors from patients treated with tions in PIK3R1 and tumor-suppressor genes. In patients BRAFiÆMEKi underwent gene expression profiling and treated with anti–PD-1, V600K (n ¼ 19) had superior DNA sequencing. Molecular results were validated using outcomes than V600E (n ¼ 84), including response rate The Cancer Genome Atlas (TCGA) data. An independent (53% vs. 29%, P ¼ 0.059), PFS (median, 19 vs. 2.7 months, cohort of V600E/K patients treated with anti–PD-1 immu- P ¼ 0.049), and overall survival (20.4 vs. -

Subtype-Dependent Difference of Glucose Transporter 1 And
www.nature.com/scientificreports OPEN Subtype‑dependent diference of glucose transporter 1 and hexokinase II expression in craniopharyngioma: an immunohistochemical study Naoto Mukada1, Masahiko Tosaka1*, Nozomi Matsumura2, Rei Yamaguchi1, Masanori Aihara1, Koji Isoda2, Tetsuya Higuchi3, Yoshito Tsushima3, Hideaki Yokoo2 & Yuhei Yoshimoto1 Papillary craniopharyngiomas are characterized by the BRAF V600E mutation. Enhancement of glucose metabolism may be involved in the downstream of the BRAF V600E mutation in many types of tumors. Glucose metabolism was investigated in craniopharyngioma using immunohistochemical analysis. The study included 29 cases of craniopharyngioma (18 adamantinomatous type [ACP], 11 papillary type [PCP]). Immunohistochemical analysis was performed with anti‑glucose transporter‑1 (GLUT‑1), anti‑hexokinase‑II (HK‑II), anti‑BRAF V600E, and anti‑beta‑catenin antibodies. Expressions of GLUT‑1 and HK‑II were evaluated using a semiquantitative 4‑tiered scale as 0, 1+, 2+, 3+, and divided into negative (0 or 1+) or positive (2+ or 3+) group. GLUT‑1 expression level was signifcantly higher in PCPs than ACPs (0, 1+, 2+, 3+ = 2, 12, 4, 0 cases in ACP, respectively, 0, 1+, 2+, 3+ = 0, 2, 5, 4 in PCP, p = 0.001), and most PCPs were classifed into positive group (positive rate, 22.2% [4/18] in ACP, 81.8% [9/11] in PCP; p = 0.003). HK‑II expression was also conspicuous in PCPs (0, 1+, 2+, 3+ = 7, 9, 2, 0 cases in ACP, 0, 3, 3, 5 in PCP; p = 0.001), and most of them divided into positive group (positive rate, 11.1% [2/18] in ACP, 72.7% [8/11] in PCP; p = 0.001). -
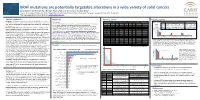
BRAF Mutations Are Potentially Targetable Alterations in a Wide
BRAF mutations are potentially targetable alterations in a wide variety of solid cancers Zoran Gatalica1*, Ken Burnett1, Ryan Bender1, Rebecca Feldman1, Semir Vranic2, Sandeep Reddy1 1 Caris Life Sciences, Phoenix, AZ, USA, 2Department of Pathology, Clinical Center, University of Sarajevo, Sarajevo, Bosnia and Herzegovina *Corresponding author: Zoran Gatalica, [email protected] Abstract (updated) Methods Results, contd. Results, contd. Background: The BRAF gene mutations are potentially targetable with several Tissue samples Figure 3. Potential therapeutically relevant alterations that co-occur with BRAF treatment modalities approved in a limited number of cancer types. The present Table 1. Frequency of V600 Mutations across Tumor Types V600_ PIK3CA pathway RAS pathway A cohort of 36,312 solid tumors were profiled at Caris Life Sciences, (Phoenix, AZ) using Sanger Histoytpe V600E V600K V600R V600E(2) V600D V600M V600_ K601delinsE study explored the spectrum and frequencies of BRAF mutations in a wide variety W604delinsR BRAF Status EGFR pos. PTEN loss AKT PIK3CA PTEN STK11 KRAS HRAS NRAS V600E 58% (148/255) 49% (536/1046) 2% (8/532) 8% (50/653) 6% (31/509) 2% (9/521) 0.2% (2/927) 0.4% (2/465) 1% (4/705) and NGS assays. Next-generation sequencing (NGS) and Sanger sequencing: CRC 578 - - - - - 1 - of solid tumors. other V600 17% (5/30) 44% (55/98) 1% (1/76) 1% (1/90) 6% (4/68) 1% (1/71) 0% (0/89) 0% (0/65) 0% (0/90) NGS was performed on enriched tumor genomic DNA isolated from formalin-fixed paraffin Melanoma 462 82 12 3 1 1 3 - non-V600 42% (84/201) 35% (246/378) 1% (5/337) 8% (28/367) 7% (21/322) 5% (15/323) 19% (73/393) 2% (6/303) 5% (20/369) Materials and Methods: 36,312 solid tumors were profiled using different gene embedded samples using the Illumina MiSeq platform. -

Efficacy of Dabrafenib Plus Trametinib Combination in Patients
cancers Brief Report Efficacy of Dabrafenib Plus Trametinib Combination in Patients with BRAF V600E-Mutant NSCLC in Real-World Setting: GFPC 01-2019 Jean-Bernard Auliac 1,* , Sophie Bayle 2, Pascal Do 3, Gwenaëlle Le Garff 4, Magali Roa 5, Lionel Falchero 6, Eric Huchot 7 , Gilles Quéré 8 , Gaëlle Jeannin 9, Anne-Cécile Métivier 10, Joëlle Hobeika 1, Florian Guisier 11 and Christos Chouaid 1 1 Department of Pneumology, CHI de Créteil, 94000 Créteil, France; [email protected] (J.H.); [email protected] (C.C.) 2 Department of Pneumology, CHU de Saint-Etienne, 42055 Saint-Etienne, France; [email protected] 3 Department of Pneumology, Centre Baclesse, 14076 Caen, France; [email protected] 4 Department of Pneumology, CH de Saint Brieuc, 22027 Saint-Brieuc, France; gwenaelle.legarff@ch-stbrieuc.fr 5 Department of Pneumology, CHI de de Fréjus Saint Raphael, 83600 Fréjus, France; [email protected] 6 Department of Pneumology, Hôpital Nord Ouest de Villefranche-sur-Saône, 69400 Gleizé, France; [email protected] 7 Department of Pneumology, CHU Réunion Site Sud, 97448 Saint-Pierre, France; [email protected] 8 Department of Pneumology, CHU de Brest, 29200 Brest, France; [email protected] 9 Department of Pneumology, CHU de Clermont-Ferrand, 63000 Clermont-Ferrand, France; [email protected] 10 Department of Pneumology, Hôpital Foch, 92150 Suresnes, France; [email protected] 11 Department of Pneumology, CHU Charles Nicolle, 76000 Rouen, France; fl[email protected] * Correspondence: [email protected] Received: 6 November 2020; Accepted: 30 November 2020; Published: 2 December 2020 Simple Summary: Mutations of the BRAF oncogene are reported in tumors of patients with non-small-cell lung cancer in 2–4% of cases (about half of them are V600E mutation).