The Indian Origin of Paternal Haplogroup R1a1* Substantiates the Autochthonous Origin of Brahmins and the Caste System
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Germanic Origins from the Perspective of the Y-Chromosome
Germanic Origins from the Perspective of the Y-Chromosome By Michael Robert St. Clair A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor in Philosophy in German in the Graduate Division of the University of California, Berkeley Committee in charge: Irmengard Rauch, Chair Thomas F. Shannon Montgomery Slatkin Spring 2012 Abstract Germanic Origins from the Perspective of the Y-Chromosome by Michael Robert St. Clair Doctor of Philosophy in German University of California, Berkeley Irmengard Rauch, Chair This dissertation holds that genetic data are a useful tool for evaluating contemporary models of Germanic origins. The Germanic languages are a branch of the Indo-European language family and include among their major contemporary representatives English, German, Dutch, Danish, Swedish, Norwegian and Icelandic. Historically, the search for Germanic origins has sought to determine where the Germanic languages evolved, and why the Germanic languages are similar to and different from other European languages. Both archaeological and linguist approaches have been employed in this research direction. The linguistic approach to Germanic origins is split among those who favor the Stammbaum theory and those favoring language contact theory. Stammbaum theory posits that Proto-Germanic separated from an ancestral Indo-European parent language. This theoretical approach accounts for similarities between Germanic and other Indo- European languages by posting a period of mutual development. Germanic innovations, on the other hand, occurred in isolation after separation from the parent language. Language contact theory posits that Proto-Germanic was the product of language convergence and this convergence explains features that Germanic shares with other Indo-European languages. -

Reassessing Religion and Politics in the Life of Jagjivan Ram¯
religions Article Reassessing Religion and Politics in the Life of Jagjivan Ram¯ Peter Friedlander South and South East Asian Studies Program, School of Culture History and Language, College of Asia and the Pacific, Australian National University, Canberra, ACT 2600, Australia; [email protected] Received: 13 March 2020; Accepted: 23 April 2020; Published: 1 May 2020 Abstract: Jagjivan Ram (1908–1986) was, for more than four decades, the leading figure from India’s Dalit communities in the Indian National Congress party. In this paper, I argue that the relationship between religion and politics in Jagjivan Ram’s career needs to be reassessed. This is because the common perception of him as a secular politician has overlooked the role that his religious beliefs played in forming his political views. Instead, I argue that his faith in a Dalit Hindu poet-saint called Ravidas¯ was fundamental to his political career. Acknowledging the role that religion played in Jagjivan Ram’s life also allows us to situate discussions of his life in the context of contemporary debates about religion and politics. Jeffrey Haynes has suggested that these often now focus on whether religion is a cause of conflict or a path to the peaceful resolution of conflict. In this paper, I examine Jagjivan Ram’s political life and his belief in the Ravidas¯ ¯ı religious tradition. Through this, I argue that Jagjivan Ram’s career shows how political and religious beliefs led to him favoring a non-confrontational approach to conflict resolution in order to promote Dalit rights. Keywords: religion; politics; India; Congress Party; Jagjivan Ram; Ravidas;¯ Ambedkar; Dalit studies; untouchable; temple building 1. -

CASTE SYSTEM in INDIA Iwaiter of Hibrarp & Information ^Titntt
CASTE SYSTEM IN INDIA A SELECT ANNOTATED BIBLIOGRAPHY Submitted in partial fulfilment of the requirements for the award of the degree of iWaiter of Hibrarp & information ^titntt 1994-95 BY AMEENA KHATOON Roll No. 94 LSM • 09 Enroiament No. V • 6409 UNDER THE SUPERVISION OF Mr. Shabahat Husaln (Chairman) DEPARTMENT OF LIBRARY & INFORMATION SCIENCE ALIGARH MUSLIM UNIVERSITY ALIGARH (INDIA) 1995 T: 2 8 K:'^ 1996 DS2675 d^ r1^ . 0-^' =^ Uo ulna J/ f —> ^^^^^^^^K CONTENTS^, • • • Acknowledgement 1 -11 • • • • Scope and Methodology III - VI Introduction 1-ls List of Subject Heading . 7i- B$' Annotated Bibliography 87 -^^^ Author Index .zm - 243 Title Index X4^-Z^t L —i ACKNOWLEDGEMENT I would like to express my sincere and earnest thanks to my teacher and supervisor Mr. Shabahat Husain (Chairman), who inspite of his many pre Qoccupat ions spared his precious time to guide and inspire me at each and every step, during the course of this investigation. His deep critical understanding of the problem helped me in compiling this bibliography. I am highly indebted to eminent teacher Mr. Hasan Zamarrud, Reader, Department of Library & Information Science, Aligarh Muslim University, Aligarh for the encourage Cment that I have always received from hijft* during the period I have ben associated with the department of Library Science. I am also highly grateful to the respect teachers of my department professor, Mohammadd Sabir Husain, Ex-Chairman, S. Mustafa Zaidi, Reader, Mr. M.A.K. Khan, Ex-Reader, Department of Library & Information Science, A.M.U., Aligarh. I also want to acknowledge Messrs. Mohd Aslam, Asif Farid, Jamal Ahmad Siddiqui, who extended their 11 full Co-operation, whenever I needed. -

Linguistic History and Language Diversity in India: Views and Counterviews
J Biosci (2019) 44:62 Indian Academy of Sciences DOI: 10.1007/s12038-019-9879-1 (0123456789().,-volV)(0123456789().,-volV) Linguistic history and language diversity in India: Views and counterviews SONAL KULKARNI-JOSHI Deccan College, Pune, India (Email, [email protected]) This paper addresses the theme of the seminar from the perspective of historical linguistics. It introduces the construct of ‘language family’ and then proceeds to a discussion of contact and the dynamics of linguistic exchange among the main language families of India over several millennia. Some prevalent hypotheses to explain the creation of India as a linguistic area are presented. The ‘substratum view’ is critically assessed. Evidence from historical linguistics in support of two dominant hypotheses –‘the Aryan migration view’ and ‘the out-of-India hypothesis’–is presented and briefly assessed. In conclusion, it is observed that the current understanding in historical linguistics favours the Aryan migration view though the ‘substratum view’ is questionable. Keywords. Aryan migration; historical linguistics; language family; Out-of-India hypothesis; substratum 1. Introduction the basis of social, political and cultural criteria more than linguistic criteria. The aim of this paper is to lend a linguistic perspective on This vast number of languages is classified into four (or the issue of human diversity and ancestry in India to the non- six) language families or genealogical types: Austro-Asiatic linguists at this seminar. The paper is an overview of the (Munda), Dravidian, Indo-Aryan (IA) and Tibeto-Burman; major views and evidences gleaned from the available more recently, two other language families have been literature. -

THE TEXTS, POLITICAL HISTORY and ADMINISTRATION Till C. 200 BC
History of Ancient India Volume III THE TEXTS, POLITICAL HISTORY AND ADMINISTRATION Till c. 200 BC Editors Dilip K. Chakrabarti and Makkhan Lal Vivekananda International Foundation New Delhi Aryan Books International New Delhi Cataloging in Publication Data—DK [Courtesy: D.K. Agencies (P) Ltd. <[email protected]>] History of ancient India / editors, Dilip K. Chakrabarti and Makkhan Lal. v. 3 cm. Contributed articles. Includes index. Contents: v. 3. The texts, political history and administration, till c. 200 BC. ISBN 9788173054822 1. India—History. 2. India—Politics and government. I. Chakrabarti, Dilip K., 1941- II. Makkhan Lal, 1954- III. Vivekananda International Foundation. DDC 954 23 ISBN: 978-81-7305-482-2 © Vivekananda International Foundation All rights reserved. No part of this book may be reproduced, utilised in any form or by any means, electronic and mechanical, including photocopying, recording or by any information storage and retrieval system without prior permission of the authors and the publishers. Responsibility for statements made and visuals provided in the various papers rest solely with the contributors. The views expressed by individual authors are not necessarily those of the editors or of publishers. First Published in 2014 by Vivekananda International Foundation 3, San Martin Marg, Chanakyapuri, New Delhi - 110 021 Tel.: 24121764, 24106698; Fax: 91-11-43115450 E-mail: [email protected] www.vifindia.org in association with Aryan Books International Pooja Apartments, 4B, Ansari Road, New Delhi - 110 002 Tel.: 23287589, 23255799; Fax: 91-11-23270385 E-mail: [email protected] www.aryanbooks.co.in Designed and Printed in India at ABI Prints & Publishing Co., New Delhi. -

R E S U M E of the Business Transacted by the Rajya Sabha 246Th Session
PARLIAMENT OF INDIA RAJYA SABHA R E S U M E OF THE BUSINESS TRANSACTED BY THE RAJYA SABHA 246TH SESSION (18th July to 10th August, 2018) Rajya Sabha Secretariat, New Delhi (Website –http://rajyasabha.nic.in) P R E F A C E This publication contains a brief resume of the business transacted by the Rajya Sabha during its Two Hundred and Forty Sixth Session. NEW DELHI; DESH DEEPAK VERMA —————— Secretary-General. September 20, 2018 C O N T E N T S PAGES 1. Summons 1 2. Duration of the Session 1 3. National Anthem 1 4. Oath or Affirmation 2 5. Obituary References 3-5 6. References by the Chair 6-7 7. Felicitations by the Chair 8 8. Papers Laid on the Table 9-13 9. Reports/Statem ents of the Committees Presented/Laid on the 14-16 Table 10. Election of Deputy Chairman 16 11. Felicitations to the Deputy Chairman 17 12. Leave of Absence 17 13. Announcements by the Chair 17-18 14. Ruling by the Chair 18-19 15. Observations by the Chair 20 16. Cancellation of Sitting of Rajya Sabha 20 17. Messages from the Lok Sabha-Reported/Government Bills 20 Laid on the Table 18. Recommendations of the Business Advisory Committee 21 19. Statements regarding Government Business 21 20. Motions f or Election of Members to various 21 Committees/Bodies 21. Motion for Extension of time for Presentation of the Report 22 of the Select Committee on the Ancient Monuments and Archaeological Sites and Remains (Amendment) Bill, 2018 22. Matters raised with permission 22-32 23. -

Y-DNA HAPLOGROUP R1A and the URALIC LANGUAGE FAMILY Csaba Barnabas Horvath Pazmany Peter Catholic University, Hungary Email: [email protected]
Second Asia Pacific Conference on Advanced Research (APCAR, Melbourne, February, 2016) ISBN:978 0 9943656 37 www.apiar.org.au Y-DNA HAPLOGROUP R1A AND THE URALIC LANGUAGE FAMILY Csaba Barnabas Horvath Pazmany Peter Catholic University, Hungary email: [email protected] Abstract My paper examines the issue of the geographic origins of the Uralic language family in the scope of the presence of Y-DNA haplo groups in the human population. The analysis of genetic statistics on the geographic and demographic distribution of Y-DNA haplo groups has recently become an innovative component in tracking down prehistoric migrations and thus, in solving the puzzle of the geographic origins of different language families. The conventional view on the geographical origins of the Uralic languages puts their origin in the subarctic regions alongside the Ural Mountains, and spread largely within the subarctic forest belt. Genetic traits, however, show evidence of large-scale Bronze Age migrations to most of the traditional Uralic language areas from the south, from the continental steppe and forest steppe belts. The mainstream approach views such events as mere incursions by Indo- Europeans. Genetic and archeological evidence, however, both indicate that these migrations reached further, both in magnitude and geographic range, than could have been plausible without the entire Uralic language family being erased and replaced by Indo-European languages. This suggests that the origins of the language family may lie in these migrations from the south, mostly marked by the dominance of the haplo group R1a. My research aims to analyze if there is a synchrony between the internal genealogical structure of the Uralic language family and such a scenario. -

Human Mtdna Hypervariable Regions, HVR I and II, Hint at Deep Common Maternal Founder and Subsequent Maternal Gene flow in Indian Population Groups
J Hum Genet (2005) 50: 497–506 DOI 10.1007/s10038-005-0284-2 ORIGINAL ARTICLE Swarkar Sharma Æ Anjana Saha Æ Ekta Rai Audesh Bhat Æ Ramesh Bamezai Human mtDNA hypervariable regions, HVR I and II, hint at deep common maternal founder and subsequent maternal gene flow in Indian population groups Received: 6 May 2005 / Accepted: 21 July 2005 / Published online: 5 October 2005 Ó The Japan Society of Human Genetics and Springer-Verlag 2005 Abstract We have analysed the hypervariable regions diversity in human populations is low relative to that in (HVR I and II) of human mitochondrial DNA many other species, attesting to the recent origin and (mtDNA) in individuals from Uttar Pradesh (UP), Bihar small size of the ancestral human population (Li and (BI) and Punjab (PUNJ), belonging to the Indo-Euro- Sadler 1991; Crouau-Roy et al 1996; Kaessmann et al pean linguistic group, and from South India (SI), that 1999). Since the seminal study of Cann et al (1987), have their linguistic roots in Dravidian language. Our mitochondrial DNA (mtDNA) data have proven to be analysis revealed the presence of known and novel extremely useful for studying human evolution, includ- mutations in both hypervariable regions in the studied ing prehistoric migrations and demographic events such population groups. Median joining network analyses as sudden population expansions or extreme bottlenecks based on mtDNA showed extensive overlap in mtDNA (Sherry et al 1994). lineages despite the extensive cultural and linguistic The human mtDNA is a closed circular genome of diversity. MDS plot analysis based on Fst distances 16.5 kb in length (Anderson et al 1981), which includes suggested increased maternal genetic proximity for the a 1.1 kb-long noncoding (control) region that represents studied population groups compared with other world a highly variable sequence (Greenberg et al 1983; Melton populations. -

Exploring the Y Chromosomal Ancestry of Modern Panamanians
RESEARCH ARTICLE Exploring the Y Chromosomal Ancestry of Modern Panamanians Viola Grugni1, Vincenza Battaglia1, Ugo Alessandro Perego2,3, Alessandro Raveane1, Hovirag Lancioni3, Anna Olivieri1, Luca Ferretti1, Scott R. Woodward2, Juan Miguel Pascale4, Richard Cooke5, Natalie Myres2,6, Jorge Motta4, Antonio Torroni1, Alessandro Achilli1,3, Ornella Semino1* 1 Department of Biology and Biotechnology, University of Pavia, Pavia, Italy, 2 Sorenson Molecular Genealogy Foundation, Salt Lake City, Utah, United States of America, 3 Department of Chemistry, Biology and Biotechnology, University of Perugia, Perugia, Italy, 4 Instituto Conmemorativo Gorgas de Estudios de la Salud, Panama City, Panama, 5 Smithsonian Tropical Research Institute, Panama City, Panama, 6 Ancestry, Provo, Utah, United States of America * [email protected] Abstract OPEN ACCESS Geologically, Panama belongs to the Central American land-bridge between North and Citation: Grugni V, Battaglia V, Perego UA, Raveane South America crossed by Homo sapiens >14 ka ago. Archaeologically, it belongs to a A, Lancioni H, Olivieri A, et al. (2015) Exploring the Y Chromosomal Ancestry of Modern Panamanians. wider Isthmo-Colombian Area. Today, seven indigenous ethnic groups account for 12.3% PLoS ONE 10(12): e0144223. doi:10.1371/journal. of Panama’s population. Five speak Chibchan languages and are characterized by low pone.0144223 genetic diversity and a high level of differentiation. In addition, no evidence of differential Editor: Francesc Calafell, Universitat Pompeu Fabra, structuring between maternally and paternally inherited genes has been reported in isth- SPAIN mian Chibchan cultural groups. Recent data have shown that 83% of the Panamanian gen- Received: March 31, 2015 eral population harbour mitochondrial DNAs (mtDNAs) of Native American ancestry. -
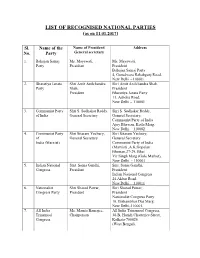
LIST of RECOGNISED NATIONAL PARTIES (As on 11.01.2017)
LIST OF RECOGNISED NATIONAL PARTIES (as on 11.01.2017) Sl. Name of the Name of President/ Address No. Party General secretary 1. Bahujan Samaj Ms. Mayawati, Ms. Mayawati, Party President President Bahujan Samaj Party 4, Gurudwara Rakabganj Road, New Delhi –110001. 2. Bharatiya Janata Shri Amit Anilchandra Shri Amit Anilchandra Shah, Party Shah, President President Bharatiya Janata Party 11, Ashoka Road, New Delhi – 110001 3. Communist Party Shri S. Sudhakar Reddy, Shri S. Sudhakar Reddy, of India General Secretary General Secretary, Communist Party of India Ajoy Bhawan, Kotla Marg, New Delhi – 110002. 4. Communist Party Shri Sitaram Yechury, Shri Sitaram Yechury, of General Secretary General Secretary India (Marxist) Communist Party of India (Marxist) ,A.K.Gopalan Bhawan,27-29, Bhai Vir Singh Marg (Gole Market), New Delhi - 110001 5. Indian National Smt. Sonia Gandhi, Smt. Sonia Gandhi, Congress President President Indian National Congress 24,Akbar Road, New Delhi – 110011 6. Nationalist Shri Sharad Pawar, Shri Sharad Pawar, Congress Party President President Nationalist Congress Party 10, Bishambhar Das Marg, New Delhi-110001. 7. All India Ms. Mamta Banerjee, All India Trinamool Congress, Trinamool Chairperson 30-B, Harish Chatterjee Street, Congress Kolkata-700026 (West Bengal). LIST OF STATE PARTIES (as on 11.01.2017) S. No. Name of the Name of President/ Address party General Secretary 1. All India Anna The General Secretary- No. 41, Kothanda Raman Dravida Munnetra in-charge Street, Chennai-600021, Kazhagam (Tamil Nadu). (Puratchi Thalaivi Amma), 2. All India Anna The General Secretary- No.5, Fourth Street, Dravida Munnetra in-charge Venkatesware Nagar, Kazhagam (Amma), Karpagam Gardens, Adayar, Chennai-600020, (Tamil Nadu). -

Urdu and the Racialized- Decastification of the “Backward Musalmaan” in India
Article CASTE: A Global Journal on Social Exclusion Vol. 1, No. 1, pp. 175–199 February 2020 brandeis.edu/j-caste ISSN 2639-4928 DOI: 10.26812/caste.v1i1.29 The Identity of Language and the Language of Erasure: Urdu and the Racialized- Decastification of the “Backward Musalmaan” in India Sanober Umar1 (Bluestone Rising Scholar Honorable Mention 2019) Abstract The decline of Urdu in post-colonial Uttar Pradesh has often been studied alongside the fall of Muslim representation in public services and the ‘job market’ in independent India. However, there remains a severe dearth in scholarship that intertwines the tropes surrounding Urdu as ‘foreign’ to India and the role that the racialization of the language played in insidiously collaborating with post-colonial governmentality which problematically ‘decastified’ and therefore circumscribed the production of ‘Muslim minority’ citizen identity. I argue that since the 1950s the polemics of Urdu and reasons cited for its lack of institutional recognition as a regional/linguistic minority language in Uttar Pradesh (until 1994) significantly informed the constitutional construction of ‘the casteless Muslim’ in the same stage setting era of the 1950s. These seemingly disparate sites of language and caste worked together to systematically deprive some of the most marginalised lower caste and Dalit Muslims access to affirmative action as their cultural-political economies witnessed a drastic fall in the early decades after Partition. This article addresses the connections between the production -

Compliance Or Defiance? the Case of Dalits and Mahadalits
Kunnath, Compliance or defiance? COMPLIANCE OR DEFIANCE? THE CASE OF DALITS AND MAHADALITS GEORGE KUNNATH Introduction Dalits, who remain at the bottom of the Indian caste hierarchy, have resisted social and economic inequalities in various ways throughout their history.1 Their struggles have sometimes taken the form of the rejection of Hinduism in favour of other religions. Some Dalit groups have formed caste-based political parties and socio-religious movements to counter upper-caste domination. These caste-based organizations have been at the forefront of mobilizing Dalit communities in securing greater benefits from the Indian state’s affirmative action programmes. In recent times, Dalit organizations have also taken to international lobbying and networking to create wider platforms for the promotion of Dalit human rights and development. Along with protest against the caste system, Dalit history is also characterized by accommodation and compliance with Brahmanical values. The everyday Dalit world is replete with stories of Dalit communities consciously or unconsciously adopting upper-caste beliefs and practices. They seem to internalize the negative images and representations of themselves and their castes that are held and propagated by the dominant groups. Dalits are also internally divided by caste, with hierarchical rankings. They themselves thus often seem to reinforce and even reproduce the same system and norms that oppress them. This article engages with both compliance and defiance by Dalit communities. Both these concepts are central to any engagement with populations living in the context of oppression and inequality. Debates in gender studies, colonial histories and subaltern studies have engaged with the simultaneous existence of these contradictory processes.