Midline Radial Glia Translocation and Corpus Callosum Formation Require FGF Signaling
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
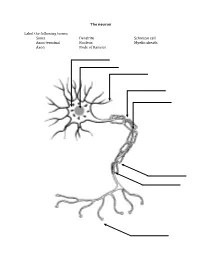
The Neuron Label the Following Terms: Soma Axon Terminal Axon Dendrite
The neuron Label the following terms: Soma Dendrite Schwaan cell Axon terminal Nucleus Myelin sheath Axon Node of Ranvier Neuron Vocabulary You must know the definitions of these terms 1. Synaptic Cleft 2. Neuron 3. Impulse 4. Sensory Neuron 5. Motor Neuron 6. Interneuron 7. Body (Soma) 8. Dendrite 9. Axon 10. Action Potential 11. Myelin Sheath (Myelin) 12. Afferent Neuron 13. Threshold 14. Neurotransmitter 15. Efferent Neurons 16. Axon Terminal 17. Stimulus 18. Refractory Period 19. Schwann 20. Nodes of Ranvier 21. Acetylcholine STEPS IN THE ACTION POTENTIAL 1. The presynaptic neuron sends neurotransmitters to postsynaptic neuron. 2. Neurotransmitters bind to receptors on the postsynaptic cell. - This action will either excite or inhibit the postsynaptic cell. - The soma becomes more positive. 3. The positive charge reaches the axon hillock. - Once the threshold of excitation is reached the neuron will fire an action potential. 4. Na+ channels open and Na+ is forced into the cell by the concentration gradient and the electrical gradient. - The neuron begins to depolarize. 5. The K+ channels open and K+ is forced out of the cell by the concentration gradient and the electrical gradient. - The neuron is depolarized. 6. The Na+ channels close at the peak of the action potential. - The neuron starts to repolarize. 7. The K+ channels close, but they close slowly and K+ leaks out. 8. The terminal buttons release neurotransmitter to the postsynaptic neuron. 9. The resting potential is overshot and the neuron falls to a -90mV (hyperpolarizes). - The neuron continues to repolarize. 10. The neuron returns to resting potential. The Synapse Label the following terms: Pre-synaptic membrane Neurotransmitters Post-synaptic membrane Synaptic cleft Vesicle Post-synaptic receptors . -

Suppression of DNA Double-Strand Break Formation by DNA Polymerase B in Active DNA Demethylation Is Required for Development of Hippocampal Pyramidal Neurons
9012 • The Journal of Neuroscience, November 18, 2020 • 40(47):9012–9027 Development/Plasticity/Repair Suppression of DNA Double-Strand Break Formation by DNA Polymerase b in Active DNA Demethylation Is Required for Development of Hippocampal Pyramidal Neurons Akiko Uyeda,1 Kohei Onishi,1 Teruyoshi Hirayama,1,2,3 Satoko Hattori,4 Tsuyoshi Miyakawa,4 Takeshi Yagi,1,2 Nobuhiko Yamamoto,1 and Noriyuki Sugo1 1Graduate School of Frontier Biosciences, Osaka University, Suita, Osaka 565-0871, Japan, 2AMED-CREST, Japan Agency for Medical Research and Development, Suita, Osaka 565-0871, Japan, 3Department of Anatomy and Developmental Neurobiology, Tokushima University Graduate School of Medical Sciences, Kuramoto, Tokushima 770-8503, Japan, and 4Institute for Comprehensive Medical Science, Fujita Health University, Toyoake, Aichi 470-1192, Japan Genome stability is essential for brain development and function, as de novo mutations during neuronal development cause psychiatric disorders. However, the contribution of DNA repair to genome stability in neurons remains elusive. Here, we demonstrate that the base excision repair protein DNA polymerase b (Polb) is involved in hippocampal pyramidal neuron fl/fl differentiation via a TET-mediated active DNA demethylation during early postnatal stages using Nex-Cre/Polb mice of ei- ther sex, in which forebrain postmitotic excitatory neurons lack Polb expression. Polb deficiency induced extensive DNA dou- ble-strand breaks (DSBs) in hippocampal pyramidal neurons, but not dentate gyrus granule cells, and to a lesser extent in neocortical neurons, during a period in which decreased levels of 5-methylcytosine and 5-hydroxymethylcytosine were observed in genomic DNA. Inhibition of the hydroxylation of 5-methylcytosine by expression of microRNAs miR-29a/b-1 diminished DSB formation. -

Crayfish Escape Behavior and Central Synapses
Crayfish Escape Behavior and Central Synapses. III. Electrical Junctions and Dendrite Spikes in Fast Flexor Motoneurons ROBERT S. ZUCKER Department of Biological Sciences and Program in the Neurological Sciences, Stanford University, Stanford, California 94305 THE LATERAL giant fiber is the decision fiber and fast flexor motoneurons are located in for a behavioral response in crayfish in- the dorsal neuropil of each abdominal gan- volving rapid tail flexion in response to glion. Dye injections and anatomical recon- abdominal tactile stimulation (27). Each structions of ‘identified motoneurons reveal lateral giant impulse is followed by a rapid that only those motoneurons which have tail flexion or tail flip, and when the giant dentritic branches in contact with a giant fiber does not fire in response to such stim- fiber are activated by that giant fiber (12, uli, the movement does not occur. 21). All of tl ie fast flexor motoneurons are The afferent limb of the reflex exciting excited by the ipsilateral giant; all but F4, the lateral giant has been described (28), F5 and F7 are also excited by the contra- and the habituation of the behavior has latkral giant (21 a). been explained in terms of the properties The excitation of these motoneurons, of this part of the neural circuit (29). seen as depolarizing potentials in their The properties of the neuromuscular somata, does not always reach threshold for junctions between the fast flexor motoneu- generating a spike which propagates out the rons and the phasic flexor muscles have also axon to the periphery (14). Indeed, only t,he been described (13). -

Plp-Positive Progenitor Cells Give Rise to Bergmann Glia in the Cerebellum
Citation: Cell Death and Disease (2013) 4, e546; doi:10.1038/cddis.2013.74 OPEN & 2013 Macmillan Publishers Limited All rights reserved 2041-4889/13 www.nature.com/cddis Olig2/Plp-positive progenitor cells give rise to Bergmann glia in the cerebellum S-H Chung1, F Guo2, P Jiang1, DE Pleasure2,3 and W Deng*,1,3,4 NG2 (nerve/glial antigen2)-expressing cells represent the largest population of postnatal progenitors in the central nervous system and have been classified as oligodendroglial progenitor cells, but the fate and function of these cells remain incompletely characterized. Previous studies have focused on characterizing these progenitors in the postnatal and adult subventricular zone and on analyzing the cellular and physiological properties of these cells in white and gray matter regions in the forebrain. In the present study, we examine the types of neural progeny generated by NG2 progenitors in the cerebellum by employing genetic fate mapping techniques using inducible Cre–Lox systems in vivo with two different mouse lines, the Plp-Cre-ERT2/Rosa26-EYFP and Olig2-Cre-ERT2/Rosa26-EYFP double-transgenic mice. Our data indicate that Olig2/Plp-positive NG2 cells display multipotential properties, primarily give rise to oligodendroglia but, surprisingly, also generate Bergmann glia, which are specialized glial cells in the cerebellum. The NG2 þ cells also give rise to astrocytes, but not neurons. In addition, we show that glutamate signaling is involved in distinct NG2 þ cell-fate/differentiation pathways and plays a role in the normal development of Bergmann glia. We also show an increase of cerebellar oligodendroglial lineage cells in response to hypoxic–ischemic injury, but the ability of NG2 þ cells to give rise to Bergmann glia and astrocytes remains unchanged. -

Oligodendrocytes in Development, Myelin Generation and Beyond
cells Review Oligodendrocytes in Development, Myelin Generation and Beyond Sarah Kuhn y, Laura Gritti y, Daniel Crooks and Yvonne Dombrowski * Wellcome-Wolfson Institute for Experimental Medicine, Queen’s University Belfast, Belfast BT9 7BL, UK; [email protected] (S.K.); [email protected] (L.G.); [email protected] (D.C.) * Correspondence: [email protected]; Tel.: +0044-28-9097-6127 These authors contributed equally. y Received: 15 October 2019; Accepted: 7 November 2019; Published: 12 November 2019 Abstract: Oligodendrocytes are the myelinating cells of the central nervous system (CNS) that are generated from oligodendrocyte progenitor cells (OPC). OPC are distributed throughout the CNS and represent a pool of migratory and proliferative adult progenitor cells that can differentiate into oligodendrocytes. The central function of oligodendrocytes is to generate myelin, which is an extended membrane from the cell that wraps tightly around axons. Due to this energy consuming process and the associated high metabolic turnover oligodendrocytes are vulnerable to cytotoxic and excitotoxic factors. Oligodendrocyte pathology is therefore evident in a range of disorders including multiple sclerosis, schizophrenia and Alzheimer’s disease. Deceased oligodendrocytes can be replenished from the adult OPC pool and lost myelin can be regenerated during remyelination, which can prevent axonal degeneration and can restore function. Cell population studies have recently identified novel immunomodulatory functions of oligodendrocytes, the implications of which, e.g., for diseases with primary oligodendrocyte pathology, are not yet clear. Here, we review the journey of oligodendrocytes from the embryonic stage to their role in homeostasis and their fate in disease. We will also discuss the most common models used to study oligodendrocytes and describe newly discovered functions of oligodendrocytes. -

Neuregulin 1–Erbb2 Signaling Is Required for the Establishment of Radial Glia and Their Transformation Into Astrocytes in Cerebral Cortex
Neuregulin 1–erbB2 signaling is required for the establishment of radial glia and their transformation into astrocytes in cerebral cortex Ralf S. Schmid*, Barbara McGrath*, Bridget E. Berechid†, Becky Boyles*, Mark Marchionni‡, Nenad Sˇ estan†, and Eva S. Anton*§ *University of North Carolina Neuroscience Center and Department of Cell and Molecular Physiology, University of North Carolina School of Medicine, Chapel Hill, NC 27599; †Department of Neurobiology, Yale University School of Medicine, New Haven, CT 06510; and ‡CeNes Pharamceuticals, Inc., Norwood, MA 02062 Communicated by Pasko Rakic, Yale University School of Medicine, New Haven, CT, January 27, 2003 (received for review December 12, 2002) Radial glial cells and astrocytes function to support the construction mine whether NRG-1-mediated signaling is involved in radial and maintenance, respectively, of the cerebral cortex. However, the glial cell development and differentiation in the cerebral cortex. mechanisms that determine how radial glial cells are established, We show that NRG-1 signaling, involving erbB2, may act in maintained, and transformed into astrocytes in the cerebral cortex are concert with Notch signaling to exert a critical influence in the not well understood. Here, we show that neuregulin-1 (NRG-1) exerts establishment, maintenance, and appropriate transformation of a critical role in the establishment of radial glial cells. Radial glial cell radial glial cells in cerebral cortex. generation is significantly impaired in NRG mutants, and this defect can be rescued by exogenous NRG-1. Down-regulation of expression Materials and Methods and activity of erbB2, a member of the NRG-1 receptor complex, leads Clonal Analysis to Study NRG’s Role in the Initial Establishment of to the transformation of radial glial cells into astrocytes. -

Chapter 8 Nervous System
Chapter 8 Nervous System I. Functions A. Sensory Input – stimuli interpreted as touch, taste, temperature, smell, sound, blood pressure, and body position. B. Integration – CNS processes sensory input and initiates responses categorizing into immediate response, memory, or ignore C. Homeostasis – maintains through sensory input and integration by stimulating or inhibiting other systems D. Mental Activity – consciousness, memory, thinking E. Control of Muscles & Glands – controls skeletal muscle and helps control/regulate smooth muscle, cardiac muscle, and glands II. Divisions of the Nervous system – 2 anatomical/main divisions A. CNS (Central Nervous System) – consists of the brain and spinal cord B. PNS (Peripheral Nervous System) – consists of ganglia and nerves outside the brain and spinal cord – has 2 subdivisions 1. Sensory Division (Afferent) – conducts action potentials from PNS toward the CNS (by way of the sensory neurons) for evaluation 2. Motor Division (Efferent) – conducts action potentials from CNS toward the PNS (by way of the motor neurons) creating a response from an effector organ – has 2 subdivisions a. Somatic Motor System – controls skeletal muscle only b. Autonomic System – controls/effects smooth muscle, cardiac muscle, and glands – 2 branches • Sympathetic – accelerator “fight or flight” • Parasympathetic – brake “resting and digesting” * 4 Types of Effector Organs: skeletal muscle, smooth muscle, cardiac muscle, and glands. III. Cells of the Nervous System A. Neurons – receive stimuli and transmit action potentials -

The Interplay Between Neurons and Glia in Synapse Development And
Available online at www.sciencedirect.com ScienceDirect The interplay between neurons and glia in synapse development and plasticity Jeff A Stogsdill and Cagla Eroglu In the brain, the formation of complex neuronal networks and regulate distinct aspects of synaptic development and amenable to experience-dependent remodeling is complicated circuit connectivity. by the diversity of neurons and synapse types. The establishment of a functional brain depends not only on The intricate communication between neurons and glia neurons, but also non-neuronal glial cells. Glia are in and their cooperative roles in synapse formation are now continuous bi-directional communication with neurons to direct coming to light due in large part to advances in genetic the formation and refinement of synaptic connectivity. This and imaging tools. This article will examine the progress article reviews important findings, which uncovered cellular made in our understanding of the role of mammalian and molecular aspects of the neuron–glia cross-talk that perisynaptic glia (astrocytes and microglia) in synapse govern the formation and remodeling of synapses and circuits. development, maturation, and plasticity since the previ- In vivo evidence demonstrating the critical interplay between ous Current Opinion article [1]. An integration of past and neurons and glia will be the major focus. Additional attention new findings of glial control of synapse development and will be given to how aberrant communication between neurons plasticity is tabulated in Box 1. and glia may contribute to neural pathologies. Address Glia control the formation of synaptic circuits Department of Cell Biology, Duke University Medical Center, Durham, In the CNS, glial cells are in tight association with NC 27710, USA synapses in all brain regions [2]. -

Neuron Morphology Influences Axon Initial Segment Plasticity1,2,3
New Research Neuronal Excitability Neuron Morphology Influences Axon Initial Segment Plasticity1,2,3 Allan T. Gulledge1 and Jaime J. Bravo2 DOI:http://dx.doi.org/10.1523/ENEURO.0085-15.2016 1Department of Physiology and Neurobiology, Geisel School of Medicine at Dartmouth, Dartmouth-Hitchcock Medical Center, Lebanon, New Hampshire 03756, and 2Thayer School of Engineering at Dartmouth, Hanover, New Hampshire 03755 Visual Abstract In most vertebrate neurons, action potentials are initiated in the axon initial segment (AIS), a specialized region of the axon containing a high density of voltage-gated sodium and potassium channels. It has recently been proposed that neurons use plasticity of AIS length and/or location to regulate their intrinsic excitability. Here we quantify the impact of neuron morphology on AIS plasticity using computational models of simplified and realistic somatodendritic morphologies. In small neurons (e.g., den- tate granule neurons), excitability was highest when the AIS was of intermediate length and located adjacent to the soma. Conversely, neu- rons having larger dendritic trees (e.g., pyramidal neurons) were most excitable when the AIS was longer and/or located away from the soma. For any given somatodendritic morphology, increasing dendritic mem- brane capacitance and/or conductance favored a longer and more distally located AIS. Overall, changes to AIS length, with corresponding changes in total sodium conductance, were far more effective in regulating neuron excitability than were changes in AIS location, while dendritic capacitance had a larger impact on AIS performance than did dendritic conductance. The somatodendritic influence on AIS performance reflects modest soma- to-AIS voltage attenuation combined with neuron size-dependent changes in AIS input resistance, effective membrane time constant, and isolation from somatodendritic capacitance. -

NERVOUS SYSTEM هذا الملف لالستزادة واثراء المعلومات Neuropsychiatry Block
NERVOUS SYSTEM هذا الملف لﻻستزادة واثراء المعلومات Neuropsychiatry block. قال تعالى: ) َو َل َق د َخ َل قنَا ا ِْلن َسا َن ِمن ُس ََل َل ة ِ من ِطي ن }12{ ثُ م َجعَ لنَاه ُ نُ ط َفة فِي َق َرا ر م ِكي ن }13{ ثُ م َخ َل قنَا ال ُّن ط َفة َ َع َل َقة َف َخ َل قنَا ا لعَ َل َقة َ ُم ضغَة َف َخ َل قنَا ا ل ُم ضغَة َ ِع َظا ما َف َك َس ونَا ا ل ِع َظا َم َل ح ما ثُ م أَن َشأنَاه ُ َخ ل قا آ َخ َر َفتَبَا َر َك ّللا ُ أَ ح َس ُن ا ل َخا ِل ِقي َن }14{( Resources BRS Embryology Book. Pathoma Book ( IN DEVELOPMENTAL ANOMALIES PART ). [email protected] 1 OVERVIEW A- Central nervous system (CNS) is formed in week 3 of development, during which time the neural plate develops. The neural plate, consisting of neuroectoderm, becomes the neural tube, which gives rise to the brain and spinal cord. B- Peripheral nervous system (PNS) is derived from three sources: 1. Neural crest cells 2. Neural tube, which gives rise to all preganglionic autonomic nerves (sympathetic and parasympathetic) and all nerves (-motoneurons and -motoneurons) that innervate skeletal muscles 3. Mesoderm, which gives rise to the dura mater and to connective tissue investments of peripheral nerve fibers (endoneurium, perineurium, and epineurium) DEVELOPMENT OF THE NEURAL TUBE Neurulation refers to the formation and closure of the neural tube. BMP-4 (bone morphogenetic protein), noggin (an inductor protein), chordin (an inductor protein), FGF-8 (fibroblast growth factor), and N-CAM (neural cell adhesion molecule) appear to play a role in neurulation. -

How Does an Axon Grow?
Downloaded from genesdev.cshlp.org on October 2, 2021 - Published by Cold Spring Harbor Laboratory Press REVIEW How does an axon grow? Jeffrey L. Goldberg1 Department of Neurobiology, Stanford University School of Medicine, Stanford, California 94305, USA How do axons grow during development, and why do cisions to advance, retract, pause, or turn (Fig. 1). All of they fail to regrow when injured? In the complicated these are potential regulatory sites that could control a mesh of our nervous system, the axon is the information neuron’s ability to elongate or regenerate its axon. superhighway, carrying all of the data we use to sense our environment and carry out behaviors. To wire up our Production of the building blocks, nervous system properly, neurons must elongate their both membrane and cytoplasmic axons during development to reach their targets. This is no simple task, however. The complex morphology of Rapid axon growth requires rapid manufacture and sup- axons and dendrites puts neurons among the most intri- ply of cytoplasm and membrane. Where are the lipid and cate and beautiful cells in the body. Knowledge of how protein building blocks made? It was known from classic neurons extend axons and dendrites, elongate at a par- experiments that axons cut off from the cell bodies of ticular rate, and stop growing at the proper time is criti- adult sensory neurons continue to elongate in culture cal to understanding the development of our nervous (Shaw and Bray 1977), but evidence for local production system, yet the regulation of these processes is poorly of membrane and cytoplasmic elements went lacking for understood. -

Regulation of Myelin Structure and Conduction Velocity by Perinodal Astrocytes
Correction NEUROSCIENCE Correction for “Regulation of myelin structure and conduc- tion velocity by perinodal astrocytes,” by Dipankar J. Dutta, Dong Ho Woo, Philip R. Lee, Sinisa Pajevic, Olena Bukalo, William C. Huffman, Hiroaki Wake, Peter J. Basser, Shahriar SheikhBahaei, Vanja Lazarevic, Jeffrey C. Smith, and R. Douglas Fields, which was first published October 29, 2018; 10.1073/ pnas.1811013115 (Proc. Natl. Acad. Sci. U.S.A. 115,11832–11837). The authors note that the following statement should be added to the Acknowledgments: “We acknowledge Dr. Hae Ung Lee for preliminary experiments that informed the ultimate experimental approach.” Published under the PNAS license. Published online June 10, 2019. www.pnas.org/cgi/doi/10.1073/pnas.1908361116 12574 | PNAS | June 18, 2019 | vol. 116 | no. 25 www.pnas.org Downloaded by guest on October 2, 2021 Regulation of myelin structure and conduction velocity by perinodal astrocytes Dipankar J. Duttaa,b, Dong Ho Wooa, Philip R. Leea, Sinisa Pajevicc, Olena Bukaloa, William C. Huffmana, Hiroaki Wakea, Peter J. Basserd, Shahriar SheikhBahaeie, Vanja Lazarevicf, Jeffrey C. Smithe, and R. Douglas Fieldsa,1 aSection on Nervous System Development and Plasticity, The Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD 20892; bThe Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., Bethesda, MD 20817; cMathematical and Statistical Computing Laboratory, Office of Intramural Research, Center for Information