Update on Blood Group Terminology and Molecular Polymorphisms
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Other Blood Group Systems—Diego,Yt, Xg, Scianna, Dombrock
Review: other blood group systems—Diego, Yt, Xg, Scianna, Dombrock, Colton, Landsteiner- Wiener, and Indian K.M. B YRNE AND P.C. B YRNE Introduction Diego Blood Group System This review was prepared to provide a basic The Diego blood group system (ISBT: DI/010) has overview of “Other Blood Groups.” Some of the more expanded from its humble beginnings to now include major blood group systems, i.e., ABO, Rh, Kell, Duffy, up to 21 discrete antigens (Table 1). 3 Band 3, an anion and Kidd, are also reviewed in this issue and are not exchange, multi-pass membrane glycoprotein, is the covered here. The sheer mass of data on the MNS basic structure that carries the Diego system antigenic blood group system is so extensive and complicated determinants. 4 The gene that encodes the Band 3 that it justifies a review all of its own, and it is therefore protein is named SLC4A1 and its chromosomal location not discussed in this article. However, various aspects is 17q12–q21. 4 of MNS were described in recent papers in Di a and Di b are antithetical, resulting from a single Immunohematology. 1,2 nucleotide substitution (2561T>C) that gives rise to The blood group systems that are covered are those amino acid changes in the Band 3 protein (Leu854Pro). that most workers believe to have some degree of To date, the Di(a–b–) phenotype has not been clinical importance or interesting features: Diego (DI), described. The Di a and Di b antigens are resistant to Yt (YT), Xg (XG), Scianna (SC), Dombrock (DO), Colton treatment with the following enzymes/chemicals: (CO), Landsteiner-Wiener (LW), and Indian (IN). -

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Viewed Under 23 (B) Or 203 (C) fi M M Male Cko Mice, and Largely Unaffected Magni Cation; Scale Bars, 500 M (B) and 50 M (C)
BRIEF COMMUNICATION www.jasn.org Renal Fanconi Syndrome and Hypophosphatemic Rickets in the Absence of Xenotropic and Polytropic Retroviral Receptor in the Nephron Camille Ansermet,* Matthias B. Moor,* Gabriel Centeno,* Muriel Auberson,* † † ‡ Dorothy Zhang Hu, Roland Baron, Svetlana Nikolaeva,* Barbara Haenzi,* | Natalya Katanaeva,* Ivan Gautschi,* Vladimir Katanaev,*§ Samuel Rotman, Robert Koesters,¶ †† Laurent Schild,* Sylvain Pradervand,** Olivier Bonny,* and Dmitri Firsov* BRIEF COMMUNICATION *Department of Pharmacology and Toxicology and **Genomic Technologies Facility, University of Lausanne, Lausanne, Switzerland; †Department of Oral Medicine, Infection, and Immunity, Harvard School of Dental Medicine, Boston, Massachusetts; ‡Institute of Evolutionary Physiology and Biochemistry, St. Petersburg, Russia; §School of Biomedicine, Far Eastern Federal University, Vladivostok, Russia; |Services of Pathology and ††Nephrology, Department of Medicine, University Hospital of Lausanne, Lausanne, Switzerland; and ¶Université Pierre et Marie Curie, Paris, France ABSTRACT Tight control of extracellular and intracellular inorganic phosphate (Pi) levels is crit- leaves.4 Most recently, Legati et al. have ical to most biochemical and physiologic processes. Urinary Pi is freely filtered at the shown an association between genetic kidney glomerulus and is reabsorbed in the renal tubule by the action of the apical polymorphisms in Xpr1 and primary fa- sodium-dependent phosphate transporters, NaPi-IIa/NaPi-IIc/Pit2. However, the milial brain calcification disorder.5 How- molecular identity of the protein(s) participating in the basolateral Pi efflux remains ever, the role of XPR1 in the maintenance unknown. Evidence has suggested that xenotropic and polytropic retroviral recep- of Pi homeostasis remains unknown. Here, tor 1 (XPR1) might be involved in this process. Here, we show that conditional in- we addressed this issue in mice deficient for activation of Xpr1 in the renal tubule in mice resulted in impaired renal Pi Xpr1 in the nephron. -

Journal of Blood Group Serology and Molecular Genetics Volume 34, Number 1, 2018 CONTENTS
Journal of Blood Group Serology and Molecular Genetics VOLUME 34, N UMBER 1, 2018 This issue of Immunohematology is supported by a contribution from Grifols Diagnostics Solutions, Inc. Dedicated to advancement and education in molecular and serologic immunohematology Immunohematology Journal of Blood Group Serology and Molecular Genetics Volume 34, Number 1, 2018 CONTENTS S EROLOGIC M ETHOD R EVIEW 1 Warm autoadsorption using ZZAP F.M. Tsimba-Chitsva, A. Caballero, and B. Svatora R EVIEW 4 Proceedings from the International Society of Blood Transfusion Working Party on Immunohaematology Workshop on the Clinical Significance of Red Blood Cell Alloantibodies, Friday, September 2, 2016, Dubai A brief overview of clinical significance of blood group antibodies M.J. Gandhi, D.M. Strong, B.I. Whitaker, and E. Petrisli C A S E R EPORT 7 Management of pregnancy sensitized with anti-Inb with monocyte monolayer assay and maternal blood donation R. Shree, K.K. Ma, L.S. Er and M. Delaney R EVIEW 11 Proceedings from the International Society of Blood Transfusion Working Party on Immunohaematology Workshop on the Clinical Significance of Red Blood Cell Alloantibodies, Friday, September 2, 2016, Dubai A review of in vitro methods to predict the clinical significance of red blood cell alloantibodies S.J. Nance S EROLOGIC M ETHOD R EVIEW 16 Recovery of autologous sickle cells by hypotonic wash E. Wilson, K. Kezeor, and M. Crosby TO THE E DITOR 19 The devil is in the details: retention of recipient group A type 5 years after a successful allogeneic bone marrow transplant from a group O donor L.L.W. -

4-6 Weeks Old Female C57BL/6 Mice Obtained from Jackson Labs Were Used for Cell Isolation
Methods Mice: 4-6 weeks old female C57BL/6 mice obtained from Jackson labs were used for cell isolation. Female Foxp3-IRES-GFP reporter mice (1), backcrossed to B6/C57 background for 10 generations, were used for the isolation of naïve CD4 and naïve CD8 cells for the RNAseq experiments. The mice were housed in pathogen-free animal facility in the La Jolla Institute for Allergy and Immunology and were used according to protocols approved by the Institutional Animal Care and use Committee. Preparation of cells: Subsets of thymocytes were isolated by cell sorting as previously described (2), after cell surface staining using CD4 (GK1.5), CD8 (53-6.7), CD3ε (145- 2C11), CD24 (M1/69) (all from Biolegend). DP cells: CD4+CD8 int/hi; CD4 SP cells: CD4CD3 hi, CD24 int/lo; CD8 SP cells: CD8 int/hi CD4 CD3 hi, CD24 int/lo (Fig S2). Peripheral subsets were isolated after pooling spleen and lymph nodes. T cells were enriched by negative isolation using Dynabeads (Dynabeads untouched mouse T cells, 11413D, Invitrogen). After surface staining for CD4 (GK1.5), CD8 (53-6.7), CD62L (MEL-14), CD25 (PC61) and CD44 (IM7), naïve CD4+CD62L hiCD25-CD44lo and naïve CD8+CD62L hiCD25-CD44lo were obtained by sorting (BD FACS Aria). Additionally, for the RNAseq experiments, CD4 and CD8 naïve cells were isolated by sorting T cells from the Foxp3- IRES-GFP mice: CD4+CD62LhiCD25–CD44lo GFP(FOXP3)– and CD8+CD62LhiCD25– CD44lo GFP(FOXP3)– (antibodies were from Biolegend). In some cases, naïve CD4 cells were cultured in vitro under Th1 or Th2 polarizing conditions (3, 4). -

Genes and Human History
Genes and human history Gil McVean, Department of Statistics, Oxford Contact: [email protected] 2 3 • Where does the variation come from? • How old are the genetic differences between us? • Are these differences important? How different are our genomes? 5 Serological techniques for detecting variation Rabbit Anti-A antibodies Human A A B AB O 6 Blood group systems in humans • 28 known systems – 39 genes, 643 alleles System Genes Alleles Knops CR1 24+ ABO ABO 102 Landsteiner- ICAM4 3 Wiener Colton C4A, C4B 7+ Lewis FUT3, FUT6 14/20 Chido-rodgers AQP1 7 Lutheran LU 16 Colton DAF 10 MNS GYPA,GYPB,G 43 Diego SLC4A1 78 YPE Dombrock DO 9 OK BSG 2 Duffy FY 9 P-related A4GALT, 14/5 Gerbich GYPC 9 B3GALT3 GIL AQP3 2 RAPH-MER2 CD151 3 H/h FUT1, FUT2 27/22 Rh RHCE, RHD, 129 I GCNT2 7 RHAG Indian CD44 2 Scianna ERMAP 4 Kell KEL, XK 33/30 Xg XG, CD99 - Kidd SLC14A1 8 YT ACHE 4 http://www.bioc.aecom.yu.edu/bgmut/summary.htm 7 Protein electroporesis • Changes in mass/charge ratio resulting from amino acid substitutions in proteins can be detected Starch or agar gel -- +- + +- - - - -- - + - + +- -- Direction of travel • In humans, about 30% of all loci show polymorphism with a 6% chance of a pair of randomly drawn alleles at a locus being different Lewontin and Hubby (1966) Harris(1966) 8 The rise of DNA sequencing GATAAGACGGTGATACTCACGCGACGGGCTTGGGCGCCGACTCGTTCAGACGGTGACCCAACTTATCCGATCGACCC CGGGTCCCGATTTAGACTCGGTATCATTTCTGGTGATTATCGCCTGCAGGTTCAAGAACACGTTTGCAGCAAGAAGT GAGGGATTTTGTCAGTGATCCCAGTCTACGGAGCCAGTCACCTCTGGTAGTGAAATTTTATTCGTTCATCTTCATAT AAGTCGCAGACCGCACGATGGGGGACAGAATACTCGCACAGGAAGAACCGCGATGAACCGAGGTAACCTAACATCCT -

Flow Reagents Single Color Antibodies CD Chart
CD CHART CD N° Alternative Name CD N° Alternative Name CD N° Alternative Name Beckman Coulter Clone Beckman Coulter Clone Beckman Coulter Clone T Cells B Cells Granulocytes NK Cells Macrophages/Monocytes Platelets Erythrocytes Stem Cells Dendritic Cells Endothelial Cells Epithelial Cells T Cells B Cells Granulocytes NK Cells Macrophages/Monocytes Platelets Erythrocytes Stem Cells Dendritic Cells Endothelial Cells Epithelial Cells T Cells B Cells Granulocytes NK Cells Macrophages/Monocytes Platelets Erythrocytes Stem Cells Dendritic Cells Endothelial Cells Epithelial Cells CD1a T6, R4, HTA1 Act p n n p n n S l CD99 MIC2 gene product, E2 p p p CD223 LAG-3 (Lymphocyte activation gene 3) Act n Act p n CD1b R1 Act p n n p n n S CD99R restricted CD99 p p CD224 GGT (γ-glutamyl transferase) p p p p p p CD1c R7, M241 Act S n n p n n S l CD100 SEMA4D (semaphorin 4D) p Low p p p n n CD225 Leu13, interferon induced transmembrane protein 1 (IFITM1). p p p p p CD1d R3 Act S n n Low n n S Intest CD101 V7, P126 Act n p n p n n p CD226 DNAM-1, PTA-1 Act n Act Act Act n p n CD1e R2 n n n n S CD102 ICAM-2 (intercellular adhesion molecule-2) p p n p Folli p CD227 MUC1, mucin 1, episialin, PUM, PEM, EMA, DF3, H23 Act p CD2 T11; Tp50; sheep red blood cell (SRBC) receptor; LFA-2 p S n p n n l CD103 HML-1 (human mucosal lymphocytes antigen 1), integrin aE chain S n n n n n n n l CD228 Melanotransferrin (MT), p97 p p CD3 T3, CD3 complex p n n n n n n n n n l CD104 integrin b4 chain; TSP-1180 n n n n n n n p p CD229 Ly9, T-lymphocyte surface antigen p p n p n -

Introduction to the Rh Blood Group.Pdf
Introduction to the Rhesus Blood Group Justin R. Rhees, M.S., MLS(ASCP)CM, SBBCM University of Utah Department of Pathology Medical Laboratory Science Program Objectives 1. Describe the major Rhesus (Rh) blood group antigens in terms of biochemical structure and inheritance. 2. Describe the characteristics of Rh antibodies. 3. Translate the five major Rh antigens, genotypes, and haplotypes from Fisher-Race to Wiener nomenclature. 4. State the purpose of Fisher-Race, Wiener, Rosenfield, and ISBT nomenclatures. Background . How did this blood group get its name? . 1937 Mrs. Seno; Bellevue hospital . Unknown antibody, unrelated to ABO . Philip Levine tested her serum against 54 ABO-compatible blood samples: only 13 were compatible. Rhesus (Rh) blood group 1930s several cases of Hemolytic of the Fetus and Newborn (HDFN) published. Hemolytic transfusion reactions (HTR) were observed in ABO- compatible transfusions. In search of more blood groups, Landsteiner and Wiener immunized rabbits with the Rhesus macaque blood of the Rhesus monkeys. Rhesus (Rh) blood group 1940 Landsteiner and Wiener reported an antibody that reacted with about 85% of human red cell samples. It was supposed that anti-Rh was the specificity causing the “intragroup” incompatibilities observed. 1941 Levine found in over 90% of erythroblastosis fetalis cases, the mother was Rh-negative and the father was Rh-positive. Rhesus macaque Rhesus (Rh) blood group Human anti-Rh and animal anti- Rh are not the same. However, “Rh” was embedded into blood group antigen terminology. The -
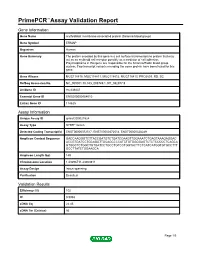
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name erythroblast membrane-associated protein (Scianna blood group) Gene Symbol ERMAP Organism Human Gene Summary The protein encoded by this gene is a cell surface transmembrane protein that may act as an erythroid cell receptor possibly as a mediator of cell adhesion. Polymorphisms in this gene are responsible for the Scianna/Radin blood group system. Two transcript variants encoding the same protein have been found for this gene. Gene Aliases MGC118810, MGC118811, MGC118812, MGC118813, PRO2801, RD, SC RefSeq Accession No. NC_000001.10, NG_008749.1, NT_032977.9 UniGene ID Hs.439437 Ensembl Gene ID ENSG00000164010 Entrez Gene ID 114625 Assay Information Unique Assay ID qHsaCID0021524 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000372517, ENST00000372514, ENST00000328249 Amplicon Context Sequence GACCAAGGGTCTTACCGATGTCTGATCCAAGTTGGAAATCTGAGTAAAGAGGAC ACCGTGATCCTGCAGGTTGCAGCCCCATCTGTGGGGAGTCTCTCCCCCTCAGCA GTGGCTCTGGCTGTGATCCTGCCTGTCCTGGTACTTCTCATCATGGTGTGCCTTT GCCTTATCTGGAAGCA Amplicon Length (bp) 149 Chromosome Location 1:43296711-43300811 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 102 R2 0.9968 cDNA Cq 22.45 cDNA Tm (Celsius) 86 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq 40.23 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report ERMAP, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference -

ERMAP Antibody (Monoclonal) (M01) Mouse Monoclonal Antibody Raised Against a Partial Recombinant ERMAP
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 ERMAP Antibody (monoclonal) (M01) Mouse monoclonal antibody raised against a partial recombinant ERMAP. Catalog # AT1943a Specification ERMAP Antibody (monoclonal) (M01) - Product Information Application WB, E Primary Accession Q96PL5 Other Accession NM_001017922 Reactivity Human Host mouse Clonality Monoclonal Isotype IgG2a Kappa Calculated MW 52605 ERMAP Antibody (monoclonal) (M01) - Additional Information Antibody Reactive Against Recombinant Protein.Western Blot detection against Gene ID 114625 Immunogen (36.74 KDa) . Other Names Erythroid membrane-associated protein, hERMAP, Radin blood group antigen, Scianna blood group antigen, ERMAP, RD, SC Target/Specificity ERMAP (NP_001017922, 376 a.a. ~ 475 a.a) partial recombinant protein with GST tag. MW of the GST tag alone is 26 KDa. Dilution WB~~1:500~1000 Format ERMAP monoclonal antibody (M01), clone Clear, colorless solution in phosphate 6F8 Western Blot analysis of ERMAP buffered saline, pH 7.2 . expression in HeLa ( (Cat # AT1943a ) Storage Store at -20°C or lower. Aliquot to avoid repeated freezing and thawing. Precautions ERMAP Antibody (monoclonal) (M01) is for research use only and not for use in diagnostic or therapeutic procedures. ERMAP Antibody (monoclonal) (M01) - Detection limit for recombinant GST tagged Page 1/2 10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 Protocols ERMAP is approximately 0.1ng/ml as a capture antibody. Provided below are standard protocols that you may find useful for product applications. ERMAP Antibody (monoclonal) (M01) - • Western Blot Background • Blocking Peptides • Dot Blot The protein encoded by this gene is a cell • Immunohistochemistry surface transmembrane protein that may act • Immunofluorescence as an erythroid cell receptor, possibly as a • Immunoprecipitation mediator of cell adhesion. -

Supp Table 6.Pdf
Supplementary Table 6. Processes associated to the 2037 SCL candidate target genes ID Symbol Entrez Gene Name Process NM_178114 AMIGO2 adhesion molecule with Ig-like domain 2 adhesion NM_033474 ARVCF armadillo repeat gene deletes in velocardiofacial syndrome adhesion NM_027060 BTBD9 BTB (POZ) domain containing 9 adhesion NM_001039149 CD226 CD226 molecule adhesion NM_010581 CD47 CD47 molecule adhesion NM_023370 CDH23 cadherin-like 23 adhesion NM_207298 CERCAM cerebral endothelial cell adhesion molecule adhesion NM_021719 CLDN15 claudin 15 adhesion NM_009902 CLDN3 claudin 3 adhesion NM_008779 CNTN3 contactin 3 (plasmacytoma associated) adhesion NM_015734 COL5A1 collagen, type V, alpha 1 adhesion NM_007803 CTTN cortactin adhesion NM_009142 CX3CL1 chemokine (C-X3-C motif) ligand 1 adhesion NM_031174 DSCAM Down syndrome cell adhesion molecule adhesion NM_145158 EMILIN2 elastin microfibril interfacer 2 adhesion NM_001081286 FAT1 FAT tumor suppressor homolog 1 (Drosophila) adhesion NM_001080814 FAT3 FAT tumor suppressor homolog 3 (Drosophila) adhesion NM_153795 FERMT3 fermitin family homolog 3 (Drosophila) adhesion NM_010494 ICAM2 intercellular adhesion molecule 2 adhesion NM_023892 ICAM4 (includes EG:3386) intercellular adhesion molecule 4 (Landsteiner-Wiener blood group)adhesion NM_001001979 MEGF10 multiple EGF-like-domains 10 adhesion NM_172522 MEGF11 multiple EGF-like-domains 11 adhesion NM_010739 MUC13 mucin 13, cell surface associated adhesion NM_013610 NINJ1 ninjurin 1 adhesion NM_016718 NINJ2 ninjurin 2 adhesion NM_172932 NLGN3 neuroligin -

Safe Blood and Blood Products
Safe Blood and Blood Products Module 3 Blood Group Serology Safe Blood and Blood Products Module 3 Blood Group Serology Conversion of electronic files for the website edition was supported by Cooperative Agreement Number PS001426 from the Centers for Disease Control and Prevention (CDC), Atlanta, United States of America. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of CDC. © World Health Organization, reprinted 2009 All rights reserved. Publications of the World Health Organization can be obtained from WHO Press, World Health Organization, 20 Avenue Appia, 1211 Geneva 27, Switzerland (tel.: +41 22 791 3264; fax: +41 22 791 4857; e-mail: [email protected]). Requests for permission to reproduce or translate WHO publications – whether for sale or for noncommercial distribution – should be addressed to WHO Press, at the above address (fax: +41 22 791 4806; e-mail: [email protected]). The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted lines on maps represent approximate border lines for which there may not yet be full agreement. The mention of specific companies or of certain manufacturers’ products does not imply that they are endorsed or recommended by the World Health Organization in preference to others of a similar nature that are not mentioned. Errors and omissions excepted, the names of proprietary products are distinguished by initial capital letters.