The Ctenophore Genome and the Evolutionary Origins of Neural Systems
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Buzzle – Zoology Terms – Glossary of Biology Terms and Definitions Http
Buzzle – Zoology Terms – Glossary of Biology Terms and Definitions http://www.buzzle.com/articles/biology-terms-glossary-of-biology-terms-and- definitions.html#ZoologyGlossary Biology is the branch of science concerned with the study of life: structure, growth, functioning and evolution of living things. This discipline of science comprises three sub-disciplines that are botany (study of plants), Zoology (study of animals) and Microbiology (study of microorganisms). This vast subject of science involves the usage of myriads of biology terms, which are essential to be comprehended correctly. People involved in the science field encounter innumerable jargons during their study, research or work. Moreover, since science is a part of everybody's life, it is something that is important to all individuals. A Abdomen: Abdomen in mammals is the portion of the body which is located below the rib cage, and in arthropods below the thorax. It is the cavity that contains stomach, intestines, etc. Abscission: Abscission is a process of shedding or separating part of an organism from the rest of it. Common examples are that of, plant parts like leaves, fruits, flowers and bark being separated from the plant. Accidental: Accidental refers to the occurrences or existence of all those species that would not be found in a particular region under normal circumstances. Acclimation: Acclimation refers to the morphological and/or physiological changes experienced by various organisms to adapt or accustom themselves to a new climate or environment. Active Transport: The movement of cellular substances like ions or molecules by traveling across the membrane, towards a higher level of concentration while consuming energy. -

J. Mar. Biol. Ass. UK (1958) 37, 7°5-752
J. mar. biol. Ass. U.K. (1958) 37, 7°5-752 Printed in Great Britain OBSERVATIONS ON LUMINESCENCE IN PELAGIC ANIMALS By J. A. C. NICOL The Plymouth Laboratory (Plate I and Text-figs. 1-19) Luminescence is very common among marine animals, and many species possess highly developed photophores or light-emitting organs. It is probable, therefore, that luminescence plays an important part in the economy of their lives. A few determinations of the spectral composition and intensity of light emitted by marine animals are available (Coblentz & Hughes, 1926; Eymers & van Schouwenburg, 1937; Clarke & Backus, 1956; Kampa & Boden, 1957; Nicol, 1957b, c, 1958a, b). More data of this kind are desirable in order to estimate the visual efficiency of luminescence, distances at which luminescence can be perceived, the contribution it makes to general back• ground illumination, etc. With such information it should be possible to discuss. more profitably such biological problems as the role of luminescence in intraspecific signalling, sex recognition, swarming, and attraction or re• pulsion between species. As a contribution to this field I have measured the intensities of light emitted by some pelagic species of animals. Most of the work to be described in this paper was carried out during cruises of R. V. 'Sarsia' and RRS. 'Discovery II' (Marine Biological Association of the United Kingdom and National Institute of Oceanography, respectively). Collections were made at various stations in the East Atlantic between 30° N. and 48° N. The apparatus for measuring light intensities was calibrated ashore at the Plymouth Laboratory; measurements of animal light were made at sea. -

Ctenophore Relationships and Their Placement As the Sister Group to All Other Animals
ARTICLES DOI: 10.1038/s41559-017-0331-3 Ctenophore relationships and their placement as the sister group to all other animals Nathan V. Whelan 1,2*, Kevin M. Kocot3, Tatiana P. Moroz4, Krishanu Mukherjee4, Peter Williams4, Gustav Paulay5, Leonid L. Moroz 4,6* and Kenneth M. Halanych 1* Ctenophora, comprising approximately 200 described species, is an important lineage for understanding metazoan evolution and is of great ecological and economic importance. Ctenophore diversity includes species with unique colloblasts used for prey capture, smooth and striated muscles, benthic and pelagic lifestyles, and locomotion with ciliated paddles or muscular propul- sion. However, the ancestral states of traits are debated and relationships among many lineages are unresolved. Here, using 27 newly sequenced ctenophore transcriptomes, publicly available data and methods to control systematic error, we establish the placement of Ctenophora as the sister group to all other animals and refine the phylogenetic relationships within ctenophores. Molecular clock analyses suggest modern ctenophore diversity originated approximately 350 million years ago ± 88 million years, conflicting with previous hypotheses, which suggest it originated approximately 65 million years ago. We recover Euplokamis dunlapae—a species with striated muscles—as the sister lineage to other sampled ctenophores. Ancestral state reconstruction shows that the most recent common ancestor of extant ctenophores was pelagic, possessed tentacles, was bio- luminescent and did not have separate sexes. Our results imply at least two transitions from a pelagic to benthic lifestyle within Ctenophora, suggesting that such transitions were more common in animal diversification than previously thought. tenophores, or comb jellies, have successfully colonized from species across most of the known phylogenetic diversity of nearly every marine environment and can be key species in Ctenophora. -

THE CASE AGAINST Marine Mammals in Captivity Authors: Naomi A
s l a m m a y t T i M S N v I i A e G t A n i p E S r a A C a C E H n T M i THE CASE AGAINST Marine Mammals in Captivity The Humane Society of the United State s/ World Society for the Protection of Animals 2009 1 1 1 2 0 A M , n o t s o g B r o . 1 a 0 s 2 u - e a t i p s u S w , t e e r t S h t u o S 9 8 THE CASE AGAINST Marine Mammals in Captivity Authors: Naomi A. Rose, E.C.M. Parsons, and Richard Farinato, 4th edition Editors: Naomi A. Rose and Debra Firmani, 4th edition ©2009 The Humane Society of the United States and the World Society for the Protection of Animals. All rights reserved. ©2008 The HSUS. All rights reserved. Printed on recycled paper, acid free and elemental chlorine free, with soy-based ink. Cover: ©iStockphoto.com/Ying Ying Wong Overview n the debate over marine mammals in captivity, the of the natural environment. The truth is that marine mammals have evolved physically and behaviorally to survive these rigors. public display industry maintains that marine mammal For example, nearly every kind of marine mammal, from sea lion Iexhibits serve a valuable conservation function, people to dolphin, travels large distances daily in a search for food. In learn important information from seeing live animals, and captivity, natural feeding and foraging patterns are completely lost. -

The Polyp and the Medusa Life on the Move
The Polyp and the Medusa Life on the Move Millions of years ago, unlikely pioneers sparked a revolution. Cnidarians set animal life in motion. So much of what we take for granted today began with Cnidarians. FROM SHAPE OF LIFE The Polyp and the Medusa Life on the Move Take a moment to follow these instructions: Raise your right hand in front of your eyes. Make a fist. Make the peace sign with your first and second fingers. Make a fist again. Open your hand. Read the next paragraph. What you just did was exhibit a trait we associate with all animals, a trait called, quite simply, movement. And not only did you just move your hand, but you moved it after passing the idea of movement through your brain and nerve cells to command the muscles in your hand to obey. To do this, your body needs muscles to move and nerves to transmit and coordinate movement, whether voluntary or involuntary. The bit of business involved in making fists and peace signs is pretty complex behavior, but it pales by comparison with the suites of thought and movement associated with throwing a curve ball, walking, swimming, dancing, breathing, landing an airplane, running down prey, or fleeing a predator. But whether by thought or instinct, you and all animals except sponges have the ability to move and to carry out complex sequences of movement called behavior. In fact, movement is such a basic part of being an animal that we tend to define animalness as having the ability to move and behave. -

University of Copenhagen, Zoological Museum, Review Universitetsparken 15, DK-2100 Copenhagen, Denmark CN, 0000-0001-6898-7655 Cite This Article: Nielsen C
Early animal evolution a morphologist's view Nielsen, Claus Published in: Royal Society Open Science DOI: 10.1098/rsos.190638 Publication date: 2019 Document version Publisher's PDF, also known as Version of record Document license: CC BY Citation for published version (APA): Nielsen, C. (2019). Early animal evolution: a morphologist's view. Royal Society Open Science, 6(7), 1-10. [190638]. https://doi.org/10.1098/rsos.190638 Download date: 30. sep.. 2021 Early animal evolution: a morphologist’s view royalsocietypublishing.org/journal/rsos Claus Nielsen The Natural History Museum of Denmark, University of Copenhagen, Zoological Museum, Review Universitetsparken 15, DK-2100 Copenhagen, Denmark CN, 0000-0001-6898-7655 Cite this article: Nielsen C. 2019 Early animal evolution: a morphologist’s view. R. Soc. open sci. Two hypotheses for the early radiation of the metazoans are vividly discussed in recent phylogenomic studies, the ‘Porifera- 6: 190638. first’ hypothesis, which places the poriferans as the sister group http://dx.doi.org/10.1098/rsos.190638 of all other metazoans, and the ‘Ctenophora-first’ hypothesis, which places the ctenophores as the sister group to all other metazoans. It has been suggested that an analysis of morphological characters (including specific molecules) could Received: 5 April 2019 throw additional light on the controversy, and this is the aim of Accepted: 4 July 2019 this paper. Both hypotheses imply independent evolution of nervous systems in Planulozoa and Ctenophora. The Porifera- first hypothesis implies no homoplasies or losses of major characters. The Ctenophora-first hypothesis shows no important synapomorphies of Porifera, Planulozoa and Placozoa. It implies Subject Category: either independent evolution, in Planulozoa and Ctenophora, of Biology (whole organism) a new digestive system with a gut with extracellular digestion, which enables feeding on larger organisms, or the subsequent Subject Areas: loss of this new gut in the Poriferans (and the re-evolution of the evolution collar complex). -

Mechanistic and Structural Studies of Pannexin Channels
MECHANISTIC AND STRUCTURAL STUDIES OF PANNEXIN CHANNELS A Dissertation Presented to the Faculty of the Graduate School of Cornell University in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy by Kevin Ronald Michalski August 2018 © 2018 Kevin Ronald Michalski MECHANISTIC AND STRUCTURAL STUDIES OF PANNEXIN CHANNELS Kevin Ronald Michalski, Ph.D Cornell University 2018 Pannexin channels are a family of recently discovered membrane proteins found in nearly every tissue of the human body. These channels have been classified as large ‘pore forming’ proteins which, when activated, create a passageway through the cell membrane through which ions and molecules transit. Current literature suggests that the actual pannexin channel is formed from a hexameric arrangement of individual monomeric pannexin subunits, resulting in a central permeation pathway for conducting ions. Opening of pannexin channels can be accomplished through several mechanisms. During apoptosis, for example, cleavage of the pannexin C-terminal domain results in a constitutively open channel through which ATP is released. However, curiously, pannexins have also been known to be activated by a variety of other stimuli such as cellular depolarization, exposure to signaling ions like Ca2+ and K+, and interacting with various other membrane receptors like members of the ATP- sensing P2X and P2Y family. How can pannexin channels sense and respond to such a diverse array of stimuli, and what is the fundamental ‘gating process’ that defines channel opening? Here, we use electrophysiology to study the activation of pannexin-1 (Panx1). We used a protein chimera approach to identify that the first extracellular domain of Panx1 is critical for inhibitor action. -

Geobiological Events in the Ediacaran Period
Geobiological Events in the Ediacaran Period Shuhai Xiao Department of Geosciences, Virginia Tech, Blacksburg, VA 24061, USA NSF; NASA; PRF; NSFC; Virginia Tech Geobiology Group; CAS; UNLV; UCR; ASU; UMD; Amherst; Subcommission of Neoproterozoic Stratigraphy; 1 Goals To review biological (e.g., acanthomorphic acritarchs; animals; rangeomorphs; biomineralizing animals), chemical (e.g., carbon and sulfur isotopes, oxygenation of deep oceans), and climatic (e.g., glaciations) events in the Ediacaran Period; To discuss integration and future directions in Ediacaran geobiology; 2 Knoll and Walter, 1992 • Acanthomorphic acritarchs in early and Ediacara fauna in late Ediacaran Period; • Strong carbon isotope variations; • Varanger-Laplandian glaciation; • What has happened since 1992? 3 Age Constraints: South China (538.2±1.5 Ma) 541 Ma Cambrian Dengying Ediacaran Sinian 551.1±0.7 Ma Doushantuo 632.5±0.5 Ma 635 Ma 635.2±0.6 Ma Nantuo (Tillite) 636 ± 5Ma Cryogenian Nanhuan 654 ± 4Ma Datangpo 663±4 Ma Neoproterozoic Neoproterozoic Jiangkou Group Banxi Group 725±10 Ma Tonian Qingbaikouan 1000 Ma • South China radiometric ages: Condon et al., 2005; Hoffmann et al., 2004; Zhou et al., 2004; Bowring et al., 2007; S. Zhang et al., 2008; Q. Zhang et al., 2008; • Additional ages from Nama Group (Namibia), Conception Group (Newfoundland), and Vendian (White Sea); 4 The Ediacaran Period Ediacara fossils Cambrian 545 Ma Nama assemblage 555 Ma White Sea assemblage 565 Ma Avalon assemblage 575 Ma 585 Ma Doushantuo biota 595 Ma 605 Ma Ediacaran Period 615 Ma -

Xenacoelomorpha's Significance for Understanding Bilaterian Evolution
Available online at www.sciencedirect.com ScienceDirect Xenacoelomorpha’s significance for understanding bilaterian evolution Andreas Hejnol and Kevin Pang The Xenacoelomorpha, with its phylogenetic position as sister biology models are the fruitfly Drosophila melanogaster and group of the Nephrozoa (Protostomia + Deuterostomia), plays the nematode Caenorhabditis elegans, in which basic prin- a key-role in understanding the evolution of bilaterian cell types ciples of developmental processes have been studied in and organ systems. Current studies of the morphological and great detail. It might be because the field of evolutionary developmental diversity of this group allow us to trace the developmental biology — EvoDevo — has its origin in evolution of different organ systems within the group and to developmental biology and not evolutionary biology that reconstruct characters of the most recent common ancestor of species under investigation are often called ‘model spe- Xenacoelomorpha. The disparity of the clade shows that there cies’. Criteria for selected representative species are cannot be a single xenacoelomorph ‘model’ species and primarily the ease of access to collected material and strategic sampling is essential for understanding the evolution their ability to be cultivated in the lab [1]. In some cases, of major traits. With this strategy, fundamental insights into the a supposedly larger number of ancestral characters or a evolution of molecular mechanisms and their role in shaping dominant role in ecosystems have played an additional animal organ systems can be expected in the near future. role in selecting model species. These arguments were Address used to attract sufficient funding for genome sequencing Sars International Centre for Marine Molecular Biology, University of and developmental studies that are cost-intensive inves- Bergen, Thormøhlensgate 55, 5008 Bergen, Norway tigations. -

GPCR Regulation of ATP Efflux from Astrocytes
G PROTEIN-COUPLED RECEPTOR REGULATION OF ATP RELEASE FROM ASTROCYTES by ANDREW EDWARD BLUM Thesis advisor: Dr. George R. Dubyak Submitted in partial fulfillment of the requirements For the degree of Doctor of Philosophy Department of Physiology and Biophysics CASE WESTERN RESERVE UNIVERSITY May, 2010 CASE WESTERN RESERVE UNIVERSITY SCHOOL OF GRADUATE STUDIES We hereby approve the thesis/dissertation of _____________________________________________________ candidate for the ______________________degree *. (signed)_______________________________________________ (chair of the committee) ________________________________________________ ________________________________________________ ________________________________________________ ________________________________________________ ________________________________________________ (date) _______________________ *We also certify that written approval has been obtained for any proprietary material contained therein. Dedication I am greatly indebted to my thesis advisor Dr. George Dubyak. Without his support, patience, and advice this work would not have been possible. I would also like to acknowledge Dr. Robert Schleimer and Dr. Walter Hubbard for their encouragement as I began my research career. My current and past thesis committee members Dr. Matthias Buck, Dr. Cathleen Carlin, Dr. Edward Greenfield, Dr. Ulrich Hopfer, Dr. Gary Landreth, Dr. Corey Smith, Dr. Jerry Silver have provided invaluable guidance and advice for which I am very grateful. A special thanks to all of the past and present -

Carbonic Anhydrases: an Ancient Tool in Calcareous Sponge Biomineralization
fgene-12-624533 March 30, 2021 Time: 13:30 # 1 BRIEF RESEARCH REPORT published: 07 April 2021 doi: 10.3389/fgene.2021.624533 Carbonic Anhydrases: An Ancient Tool in Calcareous Sponge Biomineralization Oliver Voigt1*, Benedetta Fradusco1, Carolin Gut1, Charalampos Kevrekidis1, Sergio Vargas1 and Gert Wörheide1,2,3 1 Department of Earth and Environmental Sciences, Palaeontology and Geobiology, Ludwig-Maximilians-Universität München, Munich, Germany, 2 GeoBio-Center, Ludwig-Maximilians-Universität München, Munich, Germany, 3 SNSB-Bayerische Staatssammlung für Paläontologie und Geologie, Munich, Germany Enzymes of the a-carbonic anhydrase gene family (CAs) are essential for the deposition of calcium carbonate biominerals. In calcareous sponges (phylum Porifera, class Calcarea), specific CAs are involved in the formation of calcite spicules, a unique trait and synapomorphy of this class. However, detailed studies on the CA repertoire of calcareous sponges exist for only two species of one of the two Calcarea subclasses, the Calcaronea. The CA repertoire of the second subclass, the Calcinea, has not been investigated so far, leaving a considerable gap in our knowledge about this Edited by: Melanie Debiais-Thibaud, gene family in Calcarea. Here, using transcriptomic analysis, phylogenetics, and in situ Université de Montpellier, France hybridization, we study the CA repertoire of four additional species of calcareous Reviewed by: sponges, including three from the previously unsampled subclass Calcinea. Our data Helena Cetkovi´ c,´ Rudjer Boskovic Institute, Croatia indicate that the last common ancestor of Calcarea had four ancestral CAs with defined Ana Riesgo, subcellular localizations and functions (mitochondrial/cytosolic, membrane-bound, and Natural History Museum, secreted non-catalytic). The evolution of membrane-bound and secreted CAs involved United Kingdom gene duplications and losses, whereas mitochondrial/cytosolic and non-catalytic CAs *Correspondence: Oliver Voigt are evidently orthologous genes. -
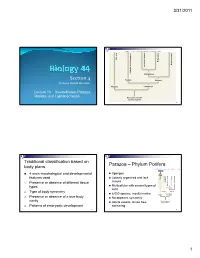
S I Section 4
3/31/2011 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Porifera Ecdysozoa Deuterostomia Lophotrochozoa Cnidaria and Ctenophora Cnidaria and Protostomia SSiection 4 Radiata Bilateria Professor Donald McFarlane Parazoa Eumetazoa Lecture 13 Invertebrates: Parazoa, Radiata, and Lophotrochozoa Ancestral colonial choanoflagellate 2 Traditional classification based on Parazoa – Phylum Porifera body plans Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Parazoa 4 main morphological and developmental Sponges features used Loosely organized and lack Porifera Ecdysozoa Cnidaria and Ctenophora tissues euterostomia photrochozoa D 1. o Presence or absence of different tissue L Multicellular with several types of types Protostomia cells 2. Type of body symmetry Radiata Bilateria 8,000 species, mostly marine Parazoa Eumetazoa 3. Presence or absence of a true body No apparent symmetry cavity Ancestral colonial Adults sessile, larvae free- choanoflagellate 4. Patterns of embryonic development swimming 3 4 1 3/31/2011 Water drawn through pores (ostia) into spongocoel Flows out through osculum Reproduce Choanocytes line spongocoel Sexually Most hermapppgggphrodites producing eggs and sperm Trap and eat small particles and plankton Gametes are derived from amoebocytes or Mesohyl between choanocytes and choanocytes epithelial cells Asexually Amoebocytes absorb food from choanocytes, Small fragment or bud may detach and form a new digest it, and carry