Current Challenges of Computational Intelligent Techniques for Functional Annotation of Ncrna Genes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Challenges Across Large-Scale Biomolecular and Polymer Simulations
Challenges across Large-Scale Biomolecular and Polymer Simulations February 21, 2017 - February 24, 2017 CECAM-AT Ivan Coluzza University of Vienna, Austria Samuela Pasquali Paris Descartes University, France Barbara Capone University of Vienna, Austria Christoph Dellago University of Vienna, Austria Tamar Schlick New York University, USA 1 Description The goal of this workshop is to bring together scientists who study large scale molecular systems both in the biological world and those from the polymer science community. This workshop aims to bring broad-minded scientists interested in these challenges to discuss state-of-the-art approaches and issues with the goal of advancing structural biophysics and applied fields. By discussing various approaches and ways to integrate knowledge on multiple scales, we hope to build a community to advance this important field. The increase in computer power in tandem with algorithmic and force-field improvements are opening opportunities for modeling large biomolecular systems as never before. Yet, many challenges in modeling and simulations of such complex systems, in realistic environments, require innovations to make the necessary experimental connections and develop new theories and mechanistic details of the associated processes. The workshop that we propose will follow and build upon the first workshop organized in Telluride (June 14-18, 2015) by Tamar Schlick and Klaus Schulten entitled "Challenges in Simulating Large-Scale Biomolecular Complexes", by focusing on the following subjects: - Large scale simulations with respect to time and system size. We will include recent advances in atomistic force-fields and technological advances in algorithms and hardware that make possible simulations on the millisecond time scales for systems composed of millions of atoms, such as large solvated biomolecules. -

Study Unveils Hidden Molecular Machinery in RNA Processes 21 September 2016
Study unveils hidden molecular machinery in RNA processes 21 September 2016 In epigenetics, the environment can produce a physiological change that gets passed down from generation to generation; however, the cells' DNA sequence is never changed and remains exactly the same. Plants are ideal model systems, Sanbonmatsu pointed out, allowing the study of epigenetic memory (where one generation of cells remembers what happened to the previous generation of cells) in slow motion. For this paper, the focus was on an RNA string known as COOLAIR that controls the timing of flowering of plants in the springtime, in that it Artist's impression of a long, non-coding RNA system. senses how long a plant has been exposed to cold. Grey/blue/red indicates main long non-coding RNA. When these RNAs are turned off or removed, Green, showing a second RNA interacting with long- plants simply do not flower. noncoding RNA. Magenta ribbons and blue barrels indicated RNA-interacting proteins. Credit: Los Alamos "Flowers may seem frivolous, compared with the National Laboratory, Kara Fischer and Karissa Sanbonmatsu working structure of the rest of the plant's leaves, stems and roots, but they actually hold the keys to survival for flowering plants," Sanbonmatsu said. A special stretch of ribonucleic acid (RNA) called "In fact, the plant's reproductive success depends COOLAIR is revealing its inner structure and critically on the precise timing of flowering in the function to scientists, displaying a striking springtime. Our discovery, finding mechanical resemblance to an RNA molecular machine, substructures inside COOLAIR, is a big piece of territory previously understood to be limited to the evidence that the RNAs may be more like cells' protein factory (the 'ribosome') and not a skill machines, and they assist in this implementation of set given to mere strings of RNA. -

˜ Ü Proceedings
ᜠƧ̑॔ȵওÜΡSƼၯÙĕÿÚÈĂ The 2nd International Symposium on Large-scale Computational Science and Engineering ਫ਼Ňो̸ႏ Proceedings Ȳ ̵ 4 Nov. 8, 2012, Thursday ̵7SUએਫ਼ɝ Science Council of Japan, Tokyo, Japan ΪŮ Sponsor Science Council of Japan Japan Science and Technology Agency ȁŮ Co-sponsor ̵7̺µ˨SU (The Japan Society for Industrial and Applied Mathematics) ̵7̴SUThe Chemical Society of Japan ̵7ɝļSU (The Japan Society of Mechanical Engineers) ̵7ওÜǜSU (The Japan Society for Computational Engineering and Science) ̵7ওÜ˨ǜSU (Japan Society for Computational Methods in Engineering) ̵7ওÜ˒SೳϤ JACM (Japan Association for Computational Mechanics) ̵7ÙāćĎĞÙĉĕSU (Japan Society for Simulation Technology) ̵7̀SU (The Physical Society of Japan) ͨଦ Support Ҝ˿తğU (Society of Automotive Engineers of Japan) ÙĕÿÚÈĂ୳̸ ಶȲÕĕöćĞáͬ೯ͮЏΆ]h:ŃĝwɰBĝฎBΡS: ǜS:ǥĝ̓˵ΡS:%Ư¦ͼƯ¦ĀčãÛÓĞč:o: ̈sï¯̀o¯Āčã÷ÅÚåÑÛϧ¶È{ʢ"pΑÒΠ ¥°n®:̒{hÒ p˨̈s̋°f®¢}; 7ÙĕÿÚÈĂ:w°ʢ"ɰญΠo:ìðĝòÆÌΚǥႸ ȏ:izΡS¬āÑď:f¯hĀÑďϧ¶ॾz}¯ɰญ¶È {:Ąéč̴¨˨ĕওÜýϚ¬ÙāćĎĞÙĉĕçȷ³¯Åᆏ h:ƼȎ̅1{h¯êåùÑċÛʢ"͵¶ŝΨ{ਫ਼Ňóïčখ ਭ¶ࢌh¢};w°¬®:ϣʢ"ᆏ̉ਭĝğ:ࣽ˨ʢ" ᆏȁ೫¦p͒o®:ʢ"ᆏ¶୨l¬®̑qÒ p̋ °¯wp/˽y°¢};¢:ͽӑ̙>¦̈˨xί˕hq:w̙ ჳÕĕöćĞáÙāćĎĞÙĉĕʢ"˲ʵÒ ¶ͽӑ̓Uȁ{h ͳlh¢}; ― 3 ― ùďÒċĂ ࿔UƅŬ ÙĕÿÚÈĂએ࿇ƅŬ ʆǕ Ǟɖ̵7SUએ f̑S ˝Dž ȁŮ͵.ࢢƅŬ ǯΨ ç͂ΡSğƊҶɝǻãæʢ"Ǟൺ ΆȅΪ=ਚÅ 78 ùĎìČਫ਼Ňìð ϞUᜨÑŦ Ïͽ̴Sʢ"ôওÜiz̀ʢ" ࿇ ĀÈď þÊďƼÛêċÛøčÒ̑SŃ̴̀Sʢ"ô ˝Dž ÚĉÝ÷ āÑčůƼÕďċë̑SþĞčâĞ× ˝Dž Πญ 1ͽ൫ůƼͩÍČ÷ËčíÄ̑S ˝Dž 78]78 O£ ]787] ùĎìČਫ਼ŇᜠòÆÌ ϞUᜨİ7 ͐ȶϊÿ̑S ˝Dž ĂĞĕ Ï ÏĂᅁƼÒDzᇻ̑S ɀ˝Dž ÍČĞ× ×ĕþĕĀæůƼďÛÄċĄÛƼWʢ"ô -

FINDING FUNCTION in MYSTERY TRANSCRIPTS Little Is Known About the Function of Most Long Non-Coding Rnas
TECHNOLOGY FEATURE FINDING FUNCTION IN MYSTERY TRANSCRIPTS Little is known about the function of most long non-coding RNAs. But a suite of new tools might change that. KARISSA Y. SANBONMATSU KARISSA Y. Conceptual model of a long non-coding RNA that provides structural scaffolding (grey) while binding to proteins (magenta) using highly structured domains (green, pink and orange). BY KELLY RAE CHI because they contain repetitive stretches of code pluripotent cells that develop into a human that sequence analysers had tended to filter out. fetus: those that comprise the ‘inner cell mass’ of n 2013, a group of researchers decided to dig It was a big blind spot. Other labs had uncov- an embryo that has yet to implant in the uterus2. deeper into a human embryonic stem-cell ered early evidence of RNAs that are rich in These RNA stretches are examples of long line called H1 — and uncovered some sur- repetitive codes and important in human stem non-coding RNAs (lncRNAs) — sequences at Iprises. H1 is one of the best known stem-cell cells. As the researchers, who were based mostly least 200 bases long that do not encode proteins. lines, yet the team managed to unearth more at California’s Stanford University, examined lncRNAs are present in many different kinds than 2,000 previously uncharacterized stretches their haul, they realized that they had hit on of tissue and are often found in specific spots of RNA1. What is more, 146 of those were exclu- exactly these kinds of RNA. Among their list inside a cell. But most lack a defined function sive to human embryonic stem cells, offering of 146 RNA sequences, says team member Vit- and, until recently, were thought to be little more tantalizing leads into pluripotency — the ability torio Sebastiano, three of the most abundant — than transcriptional noise. -

Program Information
RNA 2021 The 26th Annual Meeting of the RNA Society On-line May 25–June 4, 2021 RNA 2021 On-Line The 26th Annual Meeting of the RNA Society CORRECT THE MESSAGE CHANGE A LIFETM Locanabio’s CORRECTXTM platform is pioneering a new class of gene therapies by correcting the th th May 25 – June 4 , 2021 dysfunctional RNA that causes a broad range of neurodegenerative, neuromuscular and retinal diseases Gene Yeo – University of California San Diego, USA Katrin Karbstein – Scripps Research Institute, Florida, USA V. Narry Kim – Seoul National University, South Korea Anna Marie Pyle – Yale University, USA Xavier Roca – Nanyang Technological University, Singapore Jörg Vogel – University of Würzburg, Germany RNA 2021 • On-line GENERAL INFORMATION Citation of abstracts presented during RNA 2021 On-line (in bibliographies or other) is strictly prohibited. Material should be treated as personal communication and is to be cited only with the expressed written ® consent of the author(s). The Sequel IIe System NO UNAUTHORIZED PHOTOGRAPHY OF ANY MATTER PRESENTED DURING THE ON-LINE MEETING: To encourage sharing of unpublished data at the RNA Society Annual Meeting, taking of photographs, screenshots, videos, and/or downloading or saving Reveal the functional e ects any material is strictly prohibited. of alternative splicing with USE OF SOCIAL MEDIA: The official hash tag of the 26th Annual Meeting of the RNA Society is full-length transcript sequencing #RNA2021. The organizers encourage attendees to tweet about the amazing science they experience during the meeting. However, please respect these few simple rules when using the #RNA2021 hash tag, or when talking about the meeting on Twitter and other social media platforms: 1. -

Poster Session 10: Translation 21:00 - 22:00 Friday, 29Th May, 2020 Poster
Poster Session 10: Translation 21:00 - 22:00 Friday, 29th May, 2020 Poster 66 Translational fidelity is maintained through precise aminoacyl-tRNA accommodation dynamics gated by Elongation Factor Tu Dylan Girodat1, Scott Blanchard2, Hans-Joachim Wieden3, Karissa Sanbonmatsu1 1Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, New Mexico, USA. 2Department of Structural Biology, St. Jude Children's Research Hospital, Memphis, Tennessee, USA. 3Alberta RNA Research and Training Institute, University of Lethbridge, Lethbridge, Alberta, Canada Abstract The fidelity of translation is enigmatic, as the efficiency of cognate aminoacyl(aa)-tRNA selection by the ribosome is greater than what can be predicted from Watson-Crick base-pairing between the codon in the mRNA and the anticodon in the tRNA. The complexity of this process arises from the fact that aa-tRNA selection is a multistep process aided by auxiliary proteins such as the GTPase elongation factor (EF)-Tu, responsible for delivery of aa-tRNA to the ribosome. As such, the precise structural mechanism of how the ribosome in complex with EF-Tu selects for cognate aa-tRNA remains to be fully resolved. Here, using all-atom molecular dynamics (MD) simulations, we identify subtle differences between cognate and near-cognate aa-tRNA movement into the ribosome and how conformational rearrangements of EF-Tu aid in tRNA selection. Near-cognate aa-tRNA accommodation follows an alternative trajectory, compared to cognate aa-tRNA, leading to a misaligned position within the A-site. The origins of the alternative trajectory originate from the perturbed base-pairing between the codon and anticodon of the mRNA and tRNA, respectively. -

The Fourth AICS International Symposium
The Fourth AICS International Symposium Computer and Computational Sciences for Exascale Computing December 2—3, 2013, Kobe, JAPAN Organized and Sponsored by RIKEN AICS This Symposium is a part of RIKEN Symposium Series. Supported by: RIKEN HPCI Program for Computational Life Sciences Computational Materials Science Initiative JAMSTEC: Japan Agency for Marine-Earth Science and Technology Institute of Industrial Science, the University of Tokyo Japan Atomic Energy Agency Japan Aerospace Exploration Agency Joint Institute for Computational Fundamental Science (JICFuS) Information Initiative Center, Hokkaido University Cyberscience Center, Tohoku University Center for Computational Sciences, University of Tsukuba Information Technology Center, the University of Tokyo Global Scientific Information and Computing Center, Tokyo Institute of Technology Information Technology Center, Nagoya University Academic Center for Computing and Media Studies, Kyoto University Cybermedia Center, Osaka University Research Institute for Information Technology, Kyushu University National Institute of Informatics The Fourth AICS International Symposium Computer and Computational Sciences for Exascale Computing Program Invited Lecture Program December 2 (Monday) Opening Remarks 09:30 – 09:45 Kimihiko Hirao (RIKEN Advanced Institute for Computational Science) HPC in Computational Disaster Mitigation and Reduction 09:45 – 10:25 Seismic damage simulation of buildings in urban areas based on GPU and a physics engine Xinzheng Lu 10 (Department of Civil Engineering, -

Conservation and Function of COOLAIR Long Non-Coding Rnas in Brassica Flowering Time Control
Conservation and function of COOLAIR long non-coding RNAs in Brassica flowering time control Emily Jane Hawkes Thesis submitted for the degree of Doctor of Philosophy University of East Anglia John Innes Centre September 2017 © This copy of the thesis has been supplied on condition that anyone who consults it is understood to recognise that its copyright rests with the author and that use of any information derived there from must be in accordance with UK Copyright Law. In addition, any quotation or extract must include full attribution. This page is intentionally left blank. 2 Abstract Since their discovery long non-coding RNAs (lncRNAs) have in turn been described as essential genomic regulators or as transcriptional noise. Examples of lncRNAs with experimentally-validated function are limited, with poor nucleotide sequence conservation calling apparent functionality into question. COOLAIR lncRNAs are transcribed in the antisense direction at the Arabidopsis thaliana floral repressor gene, Flowering Locus C (FLC). Previous work has revealed a role for COOLAIR antisense RNAs in regulation of the FLC protein-coding sense transcript and, consequently, flowering time. FLC homologues are wide-spread in flowering plants, but nucleotide sequence conservation of COOLAIR-specific regions is low. COOLAIR has a complex secondary structure, and covariant base-pair mutations predict strong conservation of this secondary structure across flowering plants. Syntenic transcription of COOLAIR was confirmed in vivo for several species within the family Brassicaceae, including three commercially important Brassica crops: B. rapa, B. oleracea and B. napus. COOLAIR transcription was detected from at least three of four ancient FLC clades within the latter three polyploid and paleopolyploid species. -

Workshop: an Overdue Overhaul for Network Theory Their Way
January / February 2016 UPDATE IN THIS ISSUE > Designing difficult problems 2 > Anomalous economics 2 > Online health data 3 > School for sustainability 3 > First complexity school in India 3 > Science of city skylines 3 > Two new SFI trustees 4 > Artist Joerael Elliott 4 > Upcoming SFI events 4 RESEARCH NEWS Mapping the movements of birds and beasts Be they creatures of land, sea, or air, most animal species migrate. Whales, salmon, songbirds, and butterflies, for example, all travel thousands of kilome- ters to and from breeding and feeding Partial map of the Internet. Nodes represent IP addresses. Edge (line) lengths indicate delay duration between two nodes. Node colors: dark blue .net, .ca, .us; green grounds every year. .com, .org; red .mil, .gov, .edu; yellow .jp, .cn, .tw, .au, .de; magenta .uk, .it, .pl, .fr; gold .br, .kr, .nl; white unknown. (Image: Matt Britt, Wikimedia Commons) Theory and lab experiments suggest mi- grating en masse can help animals find Workshop: An overdue overhaul for network theory their way. Creatures traveling together are thought to pool the many direction- Networks are everywhere – from social are too simple,” says Moore, who dwells at SFI workshop, Inference on Networks: al estimates of the members of a group, interactions to species feeding relationships the intersection of physics, mathematics, and Algorithms, Phase Transitions, New Models, in essence tapping into the “wisdom of to the algorithms that pull information from computer science. “They don’t capture the and New Data. Moore co-organized the the crowd.” large datasets. Because of its broad utility in rich structure of real networks like power meeting with computer scientist Aaron So goes the hypothesis, anyway. -

ABSTRACT Title of Document: BUILDING a MAP of the DYNAMIC RIBOSOME. Suna Pelin Gulay, Doctor of Philosophy, 2015 Directed By
ABSTRACT Title of Document: BUILDING A MAP OF THE DYNAMIC RIBOSOME. Suna Pelin Gulay, Doctor of Philosophy, 2015 Directed By: Professor Jonathan D. Dinman Cell Biology and Molecular Genetics Our understanding of the static structure of the 80S eukaryotic ribosome has been enhanced by the emergence of high resolution cryo-electron microscopy and crystallography data over the past 15 years. However our understanding of the dynamic nature of the ribosome has lagged. High-throughput Selective 2’-Hydroxyl Acylation analyzed by Primer Extension (hSHAPE) is easily amenable for interrogation of rRNA dynamics. Here we report an improved method of hSHAPE data analysis and apply it to translation initiation and elongation complexes of the yeast ribosome to identify the changes in rRNA flexibility that occur during these processes. Most importantly, we have obtained complete analyses of tRNA binding and intersubunit bridge dynamics, as well as overall expansion segment dynamics, as the ribosome progresses through the translation elongation cycle. The results from these analyses suggest that (1) the yeast P site tRNA binding site is a “hybrid” between the prokaryotic and mammalian P sites, (2) there may be substates of intersubunit rotation, (3) expansion segments may have roles in accommodation. We are also able to identify a network of information pathways that connect elongation factor binding sites to all tRNA binding sites, five intersubunit bridges and two expansion segments. Future directions of this project will focus on improving the visualization of our data to better reflect the highly dynamic nature of the yeast ribosome and to reveal the underlying causes of the observed rRNA flexibility changes. -

ESI Annual Report 2017
Erwin Schrödinger International Institute 2017 for Mathematics and Physics T R O EP L R A ESI ANNU Publisher: Christoph Dellago, Director, The Erwin Schrödinger International Institute for Mathematics and Physics, University of Vienna, Boltzmanngasse 9, 1090 Vienna / Austria. Editorial Office: Christoph Dellago, Beatrix Wolf. Cover-Design: steinkellner.com Photos: Österreichische Zentralbibliothek für Physik, Philipp Steinkellner. Printing: Berger, Horn. Supported by the Austrian Federal Ministry of Education, Science and Research (BMBWF) through the University of Vienna. © 2018 Erwin Schrödinger International Institute for Mathematics and Physics. www.esi.ac.at Annual Report 2017 THE INSTITUTE PURSUES ITS MISSION FACILITIES / LOCATION THROUGH A VARIETY OF PROGRAMMES THE ERWIN SCHRÖDINGER INTERNA- THEMATIC PROGRAMMES offer the TIONAL INSTITUTE FOR MATHEMATICS opportunity for a large number of AND PHYSICS (ESI), founded in 1993 scientists at all career stages to come and part of the University of Vienna together for discussions, brainstorming, since 2011, is dedicated to the ad- seminars and collaboration. They typi- vancement of scholarly research in all cally last between 4 and 12 weeks, and areas of mathematics and physics are structured to cover several topical and, in particular, to the promotion of focus areas connected by a main theme. exchange between these disciplines. A programme may also include shorter workshop-like periods. WORKSHOPS with a duration of up to THE SENIOR RESEARCH FELLOWSHIP two weeks focus on a specific scientific PROGRAMME aims at attracting topic in mathematics or physics with internationally renowned scientists to an emphasis on communication and Vienna for visits to the ESI for up to seminar style presentations. several months. -
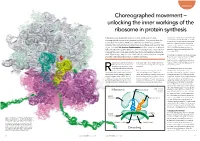
Unlocking the Inner Workings of the Ribosome in Protein Synthesis
Biophysics Choreographed movement – unlocking the inner workings of the ribosome in protein synthesis Ribosomes, tiny organelles within our cells, conduct the highly sequence for a particular protein will list the choreographed movements of protein synthesis. They ensure that the codons in the order in which the respective amino acids must be linked to one another. information encoded in DNA is successfully translated into proteins However, a ribosome cannot process the essential for nearly all chemical processes in our body, with an error rate string of codons while it is contained within of just 1 in 5000. Dr Karissa Sanbonmatsu and her team at Los Alamos DNA. The DNA must be unwound and National Laboratory, New Mexico, use cutting edge computational and copied onto a single strand called mRNA (messenger RNA). imaging tools to investigate exactly how these extraordinary structures work. Specifically, they focus on translocation, one of the most complex Ribosomes are made up of two subunits (see and little understood processes in protein synthesis. Figure 1): a small subunit where the mRNA is attached for decoding of the genetic instructions and a large subunit where the ibosomes are only 25 nanometers order specified. In other words, ribosomes respective amino acids are added to form (billionths of a meter) in diameter. convert genetic language into the language the growing protein. Despite this, they are one of the of proteins. most essential cellular organelles The mRNA strand attaches to the small involved in protein synthesis. But how does the ribosome decode the subunit of the ribosome which reads They provide the scaffold upon which the cell’s genetic information? Proteins are through each codon one at a time.