Supplementary Information 1 Family Abbreviation RTS ID Sequence
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands
The Activation of the Glucagon-Like Peptide-1 (GLP-1) Receptor by Peptide and Non-Peptide Ligands Clare Louise Wishart Submitted in accordance with the requirements for the degree of Doctor of Philosophy of Science University of Leeds School of Biomedical Sciences Faculty of Biological Sciences September 2013 I Intellectual Property and Publication Statements The candidate confirms that the work submitted is her own and that appropriate credit has been given where reference has been made to the work of others. This copy has been supplied on the understanding that it is copyright material and that no quotation from the thesis may be published without proper acknowledgement. The right of Clare Louise Wishart to be identified as Author of this work has been asserted by her in accordance with the Copyright, Designs and Patents Act 1988. © 2013 The University of Leeds and Clare Louise Wishart. II Acknowledgments Firstly I would like to offer my sincerest thanks and gratitude to my supervisor, Dr. Dan Donnelly, who has been nothing but encouraging and engaging from day one. I have thoroughly enjoyed every moment of working alongside him and learning from his guidance and wisdom. My thanks go to my academic assessor Professor Paul Milner whom I have known for several years, and during my time at the University of Leeds he has offered me invaluable advice and inspiration. Additionally I would like to thank my academic project advisor Dr. Michael Harrison for his friendship, help and advice. I would like to thank Dr. Rosalind Mann and Dr. Elsayed Nasr for welcoming me into the lab as a new PhD student and sharing their experimental techniques with me, these techniques have helped me no end in my time as a research student. -

P2Y6 Receptors Regulate CXCL10 Expression and Secretion in Mouse Intestinal Epithelial Cells
fphar-09-00149 February 26, 2018 Time: 17:57 # 1 ORIGINAL RESEARCH published: 28 February 2018 doi: 10.3389/fphar.2018.00149 P2Y6 Receptors Regulate CXCL10 Expression and Secretion in Mouse Intestinal Epithelial Cells Mabrouka Salem1,2, Alain Tremblay2, Julie Pelletier2, Bernard Robaye3 and Jean Sévigny1,2* 1 Département de Microbiologie-Infectiologie et d’Immunologie, Faculté de Médecine, Université Laval, Québec City, QC, Canada, 2 Centre de Recherche du CHU de Québec – Université Laval, Québec City, QC, Canada, 3 Institut de Recherche Interdisciplinaire en Biologie Humaine et Moléculaire, Université Libre de Bruxelles, Gosselies, Belgium In this study, we investigated the role of extracellular nucleotides in chemokine (KC, MIP- 2, MCP-1, and CXCL10) expression and secretion by murine primary intestinal epithelial cells (IECs) with a focus on P2Y6 receptors. qRT-PCR experiments showed that P2Y6 was the dominant nucleotide receptor expressed in mouse IEC. In addition, the P2Y6 Edited by: ligand UDP induced expression and secretion of CXCL10. For the other studies, we Kenneth A. Jacobson, −=− National Institutes of Health (NIH), took advantage of mice deficient in P2Y6 (P2ry6 ). Similar expression levels of P2Y1, −=− United States P2Y2, P2X2, P2X4, and A2A were detected in P2ry6 and WT IEC. Agonists of Reviewed by: TLR3 (poly(I:C)), TLR4 (LPS), P2Y1, and P2Y2 increased the expression and secretion Fernando Ochoa-Cortes, of CXCL10 more prominently in P2ry6−=− IEC than in WT IEC. CXCL10 expression Universidad Autónoma de San Luis −=− Potosí, Mexico and secretion induced by poly(I:C) in both P2ry6 and WT IEC were inhibited by Markus Neurath, general P2 antagonists (suramin and Reactive-Blue-2), by apyrase, and by specific Universitätsklinikum Erlangen, Germany antagonists of P2Y1, P2Y2, P2Y6 (only in WT), and P2X4. -

Immunosuppression Via Adenosine Receptor Activation by Adenosine
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Crossref RESEARCH ARTICLE elifesciences.org Immunosuppression via adenosine receptor activation by adenosine monophosphate released from apoptotic cells Hiroshi Yamaguchi1, Toshihiko Maruyama2, Yoshihiro Urade2†, Shigekazu Nagata1,3* 1Department of Medical Chemistry, Graduate School of Medicine, Kyoto University, Kyoto, Japan; 2Department of Molecular Behavioral Biology, Osaka Bioscience Institute, Osaka, Japan; 3Core Research for Evolutional Science and Technology, Japan Science and Technology Corporation, Kyoto, Japan Abstract Apoptosis is coupled with recruitment of macrophages for engulfment of dead cells, and with compensatory proliferation of neighboring cells. Yet, this death process is silent, and it does not cause inflammation. The molecular mechanisms underlying anti-inflammatory nature of the apoptotic process remains poorly understood. In this study, we found that the culture supernatant of apoptotic cells activated the macrophages to express anti-inflammatory genes such as Nr4a and Thbs1. A high level of AMP accumulated in the apoptotic cell supernatant in a Pannexin1-dependent manner. A nucleotidase inhibitor and A2a adenosine receptor antagonist inhibited the apoptotic *For correspondence: snagata@ supernatant-induced gene expression, suggesting AMP was metabolized to adenosine by an mfour.med.kyoto-u.ac.jp ecto-5’-nucleotidase expressed on macrophages, to activate the macrophage A2a adenosine receptor. Intraperitoneal injection of zymosan into Adora2a- or Panx1-deficient mice produced † Present address: Molecular high, sustained levels of inflammatory mediators in the peritoneal lavage. These results indicated Sleep Biology Laboratory, that AMP from apoptotic cells suppresses inflammation as a ‘calm down’ signal. WPI-International Institute for DOI: 10.7554/eLife.02172.001 Integrative Sleep Medicine, University of Tsukuba, Ibaraki, Japan Competing interests: The Introduction authors declare that no competing interests exist. -

Ligand-Gated Chloride Channels Are Receptors for Biogenic Amines in C
Ligand-Gated Chloride Channels Are Receptors for Biogenic Amines in C. elegans The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Ringstad, N., N. Abe, and H. R. Horvitz. “Ligand-Gated Chloride Channels Are Receptors for Biogenic Amines in C. elegans.” Science 325, no. 5936 (July 2, 2009): 96-100. As Published http://dx.doi.org/10.1126/science.1169243 Publisher American Association for the Advancement of Science (AAAS) Version Author's final manuscript Citable link http://hdl.handle.net/1721.1/84506 Terms of Use Creative Commons Attribution-Noncommercial-Share Alike 3.0 Detailed Terms http://creativecommons.org/licenses/by-nc-sa/3.0/ NIH Public Access Author Manuscript Science. Author manuscript; available in PMC 2010 October 25. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Science. 2009 July 3; 325(5936): 96±100. doi:10.1126/science.1169243. Ligand-gated chloride channels are receptors for biogenic amines in C. elegans Niels Ringstad1,2, Namiko Abe1,2,3, and H. Robert Horvitz1 1HHMI, Department of Biology and McGovern Institute for Brain Research, MIT, Cambridge MA 02139 Abstract Biogenic amines such as serotonin and dopamine are intercellular signaling molecules that function widely as neurotransmitters and neuromodulators. We have identified in the nematode Caenorhabditis elegans three ligand-gated chloride channels that are receptors for biogenic amines: LGC-53 is a high-affinity dopamine receptor, LGC-55 is a high-affinity tyramine receptor, and LGC-40 is a low-affinity serotonin receptor that is also gated by choline and acetylcholine. -

4695389.Pdf (3.200Mb)
Non-classical amine recognition evolved in a large clade of olfactory receptors The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Li, Qian, Yaw Tachie-Baffour, Zhikai Liu, Maude W Baldwin, Andrew C Kruse, and Stephen D Liberles. 2015. “Non-classical amine recognition evolved in a large clade of olfactory receptors.” eLife 4 (1): e10441. doi:10.7554/eLife.10441. http://dx.doi.org/10.7554/ eLife.10441. Published Version doi:10.7554/eLife.10441 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:23993622 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA RESEARCH ARTICLE Non-classical amine recognition evolved in a large clade of olfactory receptors Qian Li1, Yaw Tachie-Baffour1, Zhikai Liu1, Maude W Baldwin2, Andrew C Kruse3, Stephen D Liberles1* 1Department of Cell Biology, Harvard Medical School, Boston, United States; 2Department of Organismic and Evolutionary Biology, Museum of Comparative Zoology, Harvard University, Cambridge, United States; 3Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, United States Abstract Biogenic amines are important signaling molecules, and the structural basis for their recognition by G Protein-Coupled Receptors (GPCRs) is well understood. Amines are also potent odors, with some activating olfactory trace amine-associated receptors (TAARs). Here, we report that teleost TAARs evolved a new way to recognize amines in a non-classical orientation. -

Endothelial-Protective Effects of a G-Protein-Biased Sphingosine-1 Phosphate Receptor-1 Agonist, SAR247799, in Type-2 Diabetes R
medRxiv preprint doi: https://doi.org/10.1101/2020.05.15.20103101; this version posted May 20, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission. 1 Endothelial-protective effects of a G-protein-biased sphingosine-1 phosphate receptor-1 agonist, SAR247799, in type-2 diabetes rats and a randomized placebo-controlled patient trial. Luc Bergougnan1, Grit Andersen2, Leona Plum-Mörschel3, Maria Francesca Evaristi1, Bruno Poirier1, Agnes Tardat4, Marcel Ermer3, Theresa Herbrand2, Jorge Arrubla2, Hans Veit Coester2, Roberto Sansone5, Christian Heiss6, Olivier Vitse4, Fabrice Hurbin4, Rania Boiron1, Xavier Benain4, David Radzik1, Philip Janiak1, Anthony J Muslin7, Lionel Hovsepian1, Stephane Kirkesseli1, Paul Deutsch8, Ashfaq A Parkar8 1 Sanofi R&D, 1 Avenue Pierre Brossolette, 91385 Chilly Mazarin, France; 2 Profil Institut für Stoffwechselforschung GmbH, Hellersbergstraße 9, 41460 Neuss, Germany; 3 Profil Mainz GmbH & Co. KG, Malakoff-Passage, Rheinstraße 4C, Eingang via Templerstraße, D-55116 Mainz, Germany; 4 Sanofi R&D, 371 Rue du Professeur Blayac, 34080 Montpellier, France; 5 University Hospital Düsseldorf, Division of Cardiology, Pulmonary diseases and Vascular medicine, 40225 Düsseldorf, Germany; 6 Department of Clinical and Experimental Medicine, University of Surrey, Stag Hill, Guildford GU2 7XH, UK; 7 Sanofi US Services, 640 Memorial Drive, Cambridge MA 02139, USA; 8 Sanofi US Services, 55 Corporate Drive, Bridgewater, NJ 08807, USA. The authors confirm that the Principal Investigators for the clinical study were Grit Anderson and Leona Plum- Mörschel and that they had direct clinical responsibility for patients at the Neuss and Mainz sites, respectively. -
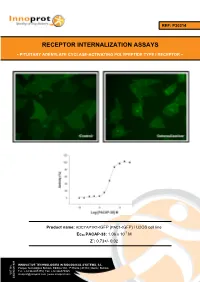
Receptor Internalization Assays
REF: P30214 RECEPTOR INTERNALIZATION ASSAYS - PITUITARY ADENYLATE CYCLASE-ACTIVATING POLYPEPTIDE TYPE I RECEPTOR - Product name: ADCYAP1R1-tGFP (PAC1-tGFP) / U2OS cell line -7 Ec50 PACAP-38: 1.06 x 10 M Z´: 0.73+/- 0.02 INNOVATIVE TECHNOLOGIES IN BIOLOGICAL SYSTEMS, S.L. Parque Tecnológico Bizkaia, Edifício 502, 1ª Planta | 48160 | Derio | Bizkaia Tel.: +34 944005355 | Fax: +34 946579925 VAT No. [email protected] | www.innoprot.com ESB95481909 Product Name: ADCYAP1R1-tGFP_U2OS Reference: P30214 Rep. Official Full Name: Pituitary adenylate cyclase- activating polypeptide type I receptor DNA Accession Number: Gene Bank AY366498 Host Cell: U2OS References: P30214: 2 vials of 3 x 106 proliferative cells P30214-DA: 1 vial of 2 x 106 division-arrested cells Storage: Liquid Nitrogen Assay Briefly description About ADCYAP1R1 Each vial of ADCYAP1R1 Internalization Assay Pituitary adenylate cyclase-activating Cell Line contains U2OS cells stably expressing polypeptide type I receptor, also known as human Pituitary adenylate cyclase-activating PAC1 is a protein that in humans is encoded by polypeptide type I receptor tagged in the N- the ADCYAP1R1 gene. ADCYAP1R1 is a terminus with tGFP protein. membrane-associated protein and shares significant homology with members of the Innoprot’s ADCYAP1R1-tGFP Internalization glucagon/secretin receptor family. This receptor Assay Cell Line has been designed to assay binds pituitary adenylate cyclase activating potential agonists/ antagonists against peptide (PACAP) mediating several biological ADCYAP1R1, modulating its activation and the activities and it is positively coupled to following redistribution process inside the cells. adenylate cyclase. This cell line will allow the image analysis of the stimuli induced by the compounds. -

Mechanisms of Acetylcholine Receptor Loss in Myasthenia Gravis
J Neurol Neurosurg Psychiatry: first published as 10.1136/jnnp.43.7.601 on 1 July 1980. Downloaded from Journal of Neurology, Neurosurgery, and Psychiatry, 1980, 43, 601-610 Mechanisms of acetylcholine receptor loss in myasthenia gravis DANIEL B DRACHMAN, ROBERT N ADAMS, ELIS F STANLEY, AND ALAN PESTRONK From the Department of Neurology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA SUMMARY The fundamental abnormality affecting the neuromuscular junctions of myasthenic patients is a reduction of available AChRs, due to an autoimmune attack directed against the receptors. Antibodies to AChR are present in most patients, and there is evidence that they have a predominant pathogenic role in the disease, aided by complement. The mechanism of antibody action involves acceleration of the rate of degradation of AChRs, attributable to cross-linking of the receptors. In addition, antibodies may block AChRs, and may participate in producing destructive changes, perhaps in conjunction with complement. The possibility that cell-mediated mechanisms may play a role in the autoimmune responses of some myasthenic patients remains to be explored. Although the target of the autoimmune attack in myasthenic patients is probably always the acetyl- Protected by copyright. choline receptors, it is not yet clear which of these immune mechanisms are most important. It is likely that the relative role of each mechanism varies from patient to patient. One of the goals of future research will be to identify the relative importance of each -

Cognition and Steroidogenesis in the Rhesus Macaque
Cognition and Steroidogenesis in the Rhesus Macaque Krystina G Sorwell A DISSERTATION Presented to the Department of Behavioral Neuroscience and the Oregon Health & Science University School of Medicine in partial fulfillment of the requirements for the degree of Doctor of Philosophy November 2013 School of Medicine Oregon Health & Science University CERTIFICATE OF APPROVAL This is to certify that the PhD dissertation of Krystina Gerette Sorwell has been approved Henryk Urbanski Mentor/Advisor Steven Kohama Member Kathleen Grant Member Cynthia Bethea Member Deb Finn Member 1 For Lily 2 TABLE OF CONTENTS Acknowledgments ......................................................................................................................................................... 4 List of Figures and Tables ............................................................................................................................................. 7 List of Abbreviations ................................................................................................................................................... 10 Abstract........................................................................................................................................................................ 13 Introduction ................................................................................................................................................................. 15 Part A: Central steroidogenesis and cognition ............................................................................................................ -

Molecular Dissection of G-Protein Coupled Receptor Signaling and Oligomerization
MOLECULAR DISSECTION OF G-PROTEIN COUPLED RECEPTOR SIGNALING AND OLIGOMERIZATION BY MICHAEL RIZZO A Dissertation Submitted to the Graduate Faculty of WAKE FOREST UNIVERSITY GRADUATE SCHOOL OF ARTS AND SCIENCES in Partial Fulfillment of the Requirements for the Degree of DOCTOR OF PHILOSOPHY Biology December, 2019 Winston-Salem, North Carolina Approved By: Erik C. Johnson, Ph.D. Advisor Wayne E. Pratt, Ph.D. Chair Pat C. Lord, Ph.D. Gloria K. Muday, Ph.D. Ke Zhang, Ph.D. ACKNOWLEDGEMENTS I would first like to thank my advisor, Dr. Erik Johnson, for his support, expertise, and leadership during my time in his lab. Without him, the work herein would not be possible. I would also like to thank the members of my committee, Dr. Gloria Muday, Dr. Ke Zhang, Dr. Wayne Pratt, and Dr. Pat Lord, for their guidance and advice that helped improve the quality of the research presented here. I would also like to thank members of the Johnson lab, both past and present, for being valuable colleagues and friends. I would especially like to thank Dr. Jason Braco, Dr. Jon Fisher, Dr. Jake Saunders, and Becky Perry, all of whom spent a great deal of time offering me advice, proofreading grants and manuscripts, and overall supporting me through the ups and downs of the research process. Finally, I would like to thank my family, both for instilling in me a passion for knowledge and education, and for their continued support. In particular, I would like to thank my wife Emerald – I am forever indebted to you for your support throughout this process, and I will never forget the sacrifices you made to help me get to where I am today. -

Protease Effects on the Structure of Acetylcholine Receptor Membranes from Torpedo Californica
PROTEASE EFFECTS ON THE STRUCTURE OF ACETYLCHOLINE RECEPTOR MEMBRANES FROM TORPEDO CALIFORNICA MICHAEL W. KLYMKOWSKY, JOHN E . HEUSER, and ROBERT M. STROUD From the Department of Biochemistry & Biophysics, University of California at San Francisco, San Francisco, California 94143 . Dr . Klymkowsky's present address is MRC Neuroimmunology Project, Department of Zoology, University College London, London WC IE, 6BT, England ABSTRACT Protease digestion of acetylcholine receptor-rich membranes derived from Torpedo californica electroplaques by homogenization and isopycnic centrifugation results in degradation of all receptor subunits without any significant effect on the appearance in electron micrographs, the toxin binding ability, or the sedimentation value of the receptor molecule . Such treatment does produce dramatic changes in the morphology of the normally 0.5- to 2-lm-diameter spherical vesicles when observed by either negative-stain or freeze-fracture electron microscopy . Removal of peripheral, apparently nonreceptor polypeptides by alkali stripping (Neubig et al ., 1979, Proc. Natl. Acad. Sci. U. S. A. 76:690-694) results in increased sensitivity of the acetylcholine receptor membranes to the protease trypsin as indicated by SDS gel electrophoretic patterns and by the extent of morphologic change observed in vesicle structure . Trypsin digestion of alkali-stripped receptor membranes results in a limit degradation pattern of all four receptor subunits, whereupon all the vesicles undergo the morphological transformation to minivesicles -

Interplay Between Gating and Block of Ligand-Gated Ion Channels
brain sciences Review Interplay between Gating and Block of Ligand-Gated Ion Channels Matthew B. Phillips 1,2, Aparna Nigam 1 and Jon W. Johnson 1,2,* 1 Department of Neuroscience, University of Pittsburgh, Pittsburgh, PA 15260, USA; [email protected] (M.B.P.); [email protected] (A.N.) 2 Center for Neuroscience, University of Pittsburgh, Pittsburgh, PA 15260, USA * Correspondence: [email protected]; Tel.: +1-(412)-624-4295 Received: 27 October 2020; Accepted: 26 November 2020; Published: 1 December 2020 Abstract: Drugs that inhibit ion channel function by binding in the channel and preventing current flow, known as channel blockers, can be used as powerful tools for analysis of channel properties. Channel blockers are used to probe both the sophisticated structure and basic biophysical properties of ion channels. Gating, the mechanism that controls the opening and closing of ion channels, can be profoundly influenced by channel blocking drugs. Channel block and gating are reciprocally connected; gating controls access of channel blockers to their binding sites, and channel-blocking drugs can have profound and diverse effects on the rates of gating transitions and on the stability of channel open and closed states. This review synthesizes knowledge of the inherent intertwining of block and gating of excitatory ligand-gated ion channels, with a focus on the utility of channel blockers as analytic probes of ionotropic glutamate receptor channel function. Keywords: ligand-gated ion channel; channel block; channel gating; nicotinic acetylcholine receptor; ionotropic glutamate receptor; AMPA receptor; kainate receptor; NMDA receptor 1. Introduction Neuronal information processing depends on the distribution and properties of the ion channels found in neuronal membranes.