Taxonomy and Phylogeny of the Brown-Rot Fungi: Fomitopsis and Its Related Genera
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
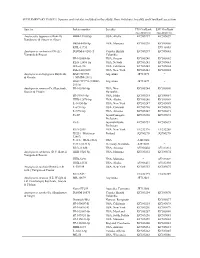
Suppl Table I
SUPPLEMENTARY TABLE I. Species and isolates included in the study, their vouchers, locality and GenBank accession Species Isolate number Locality ITS GenBank LSU GenBank accession no. accession no. Amylocystis lapponica (Romell) HHB-13400-Sp USA, Alaska KC585237 KC585059 Bondartsev & Singer ex Singer OKM-4418-Sp USA, Montana KC585238 KC585060 KHL-11755 − EU118603 Amyloporia carbonica (Overh.) DAOM-F-8281-T Canada, British KC585239 KC585061 Vampola & Pouzar Columbia FP-105585-Sp USA, Oregon KC585240 KC585062 RLG-12496-Sp USA, Nevada KC585241 KC585063 Wilcox-96 USA, California KC585242 KC585064 Zabel-40-GLN USA, New York KC585243 KC585065 Amyloporia nothofaginea Rajchenb. BAFC519794 Argentina JF713078 − & Gorjón (=MMBP-2011) BAFC519796 (=MBP- Argentina JF713079 − 2011a) Amyloporia sinuosa (Fr.) Rajchenb., FP-105386-Sp USA, New KC585244 KC585066 Gorjón & Pildain Hampshire FP-94464-Sp USA, Idaho KC585245 KC585067 HHB-12878-Sp USA, Alaska KC585246 KC585068 L-14130-Sp USA, New York KC585247 KC585069 L-6192-Sp USA, Colorado KC585248 KC585070 L-9792-Sp USA, Arizona KC585249 KC585071 Pa-3C JapanYamagata KC585250 KC585072 Prefecture Pa-3e JapanShizuoka KC585251 KC585073 Prefecture RLG-2538 USA, New York EU232196 EU232288 X725 (=Miettinen- Finland JQ700270 JQ700270 12407) P-115 (=RLG-2538) USA AJ416068 − P-211 (G-214) Germany, Karlsrube AJ345011 − RLG-1182R USA, Arizona AY966450 AY333831 Amyloporia sitchensis (D.V. Baxter) HHB-5320-Sp USA, Montana KC585252 KC585074 Vampola & Pouzar HHB-5298 USA, Montana − AY333829 HHB-12513 USA, Alaska AY966451 AY333830 -

Annotated Check List and Host Index Arizona Wood
Annotated Check List and Host Index for Arizona Wood-Rotting Fungi Item Type text; Book Authors Gilbertson, R. L.; Martin, K. J.; Lindsey, J. P. Publisher College of Agriculture, University of Arizona (Tucson, AZ) Rights Copyright © Arizona Board of Regents. The University of Arizona. Download date 28/09/2021 02:18:59 Link to Item http://hdl.handle.net/10150/602154 Annotated Check List and Host Index for Arizona Wood - Rotting Fungi Technical Bulletin 209 Agricultural Experiment Station The University of Arizona Tucson AÏfJ\fOTA TED CHECK LI5T aid HOST INDEX ford ARIZONA WOOD- ROTTlNg FUNGI /. L. GILßERTSON K.T IyIARTiN Z J. P, LINDSEY3 PRDFE550I of PLANT PATHOLOgY 2GRADUATE ASSISTANT in I?ESEARCI-4 36FZADAATE A5 S /STANT'" TEACHING Z z l'9 FR5 1974- INTRODUCTION flora similar to that of the Gulf Coast and the southeastern United States is found. Here the major tree species include hardwoods such as Arizona is characterized by a wide variety of Arizona sycamore, Arizona black walnut, oaks, ecological zones from Sonoran Desert to alpine velvet ash, Fremont cottonwood, willows, and tundra. This environmental diversity has resulted mesquite. Some conifers, including Chihuahua pine, in a rich flora of woody plants in the state. De- Apache pine, pinyons, junipers, and Arizona cypress tailed accounts of the vegetation of Arizona have also occur in association with these hardwoods. appeared in a number of publications, including Arizona fungi typical of the southeastern flora those of Benson and Darrow (1954), Nichol (1952), include Fomitopsis ulmaria, Donkia pulcherrima, Kearney and Peebles (1969), Shreve and Wiggins Tyromyces palustris, Lopharia crassa, Inonotus (1964), Lowe (1972), and Hastings et al. -

Why Mushrooms Have Evolved to Be So Promiscuous: Insights from Evolutionary and Ecological Patterns
fungal biology reviews 29 (2015) 167e178 journal homepage: www.elsevier.com/locate/fbr Review Why mushrooms have evolved to be so promiscuous: Insights from evolutionary and ecological patterns Timothy Y. JAMES* Department of Ecology and Evolutionary Biology, University of Michigan, Ann Arbor, MI 48109, USA article info abstract Article history: Agaricomycetes, the mushrooms, are considered to have a promiscuous mating system, Received 27 May 2015 because most populations have a large number of mating types. This diversity of mating Received in revised form types ensures a high outcrossing efficiency, the probability of encountering a compatible 17 October 2015 mate when mating at random, because nearly every homokaryotic genotype is compatible Accepted 23 October 2015 with every other. Here I summarize the data from mating type surveys and genetic analysis of mating type loci and ask what evolutionary and ecological factors have promoted pro- Keywords: miscuity. Outcrossing efficiency is equally high in both bipolar and tetrapolar species Genomic conflict with a median value of 0.967 in Agaricomycetes. The sessile nature of the homokaryotic Homeodomain mycelium coupled with frequent long distance dispersal could account for selection favor- Outbreeding potential ing a high outcrossing efficiency as opportunities for choosing mates may be minimal. Pheromone receptor Consistent with a role of mating type in mediating cytoplasmic-nuclear genomic conflict, Agaricomycetes have evolved away from a haploid yeast phase towards hyphal fusions that display reciprocal nuclear migration after mating rather than cytoplasmic fusion. Importantly, the evolution of this mating behavior is precisely timed with the onset of diversification of mating type alleles at the pheromone/receptor mating type loci that are known to control reciprocal nuclear migration during mating. -

Buglossoporus Pulvinus (Pers.) Donk) Входит В Число Редких Видов Макромицетов, Имеющих Высокий Природоохранный Статус Во Многих Странах Европы
УДК 582.284 : 581.527+581.42 (477.54) РЕДКИЙ ГРИБ PIPTOPORUS QUERCINUS (SCHRAD.) P. KARST. ИЗ НАЦИОНАЛЬНОГО ПРИРОДНОГО ПАРКА «ГОМОЛЬШАНСКИЕ ЛЕСА» Ордынец А.В., Акулов А.Ю. Кафедра микологии и фитоиммунологии, Харьковский национальный университет им. В.Н. Каразина пл. Свободы, 4, 61077, г. Харьков, Украина; e-mail: [email protected] Abstract. The article is devoted to a rare macromycete species Piptoporus quercinus (Schrad.) P. Karst., which has the high nature-conservative status and is included to Red lists of many European countries. The information about finding of this species on the territory of National nature park "Gomolshanskie lesa" (Zmiev area of Kharkiv district, Ukraine) is resulted. The localities of species detection on the territory of National park, the morphological, taxonomical and ecological characteristics of species, and the action plan for P. quercinus conservation are specified. Трутовый гриб Piptoporus quercinus (Schrad.) P. Karst. (=Buglossoporus pulvinus (Pers.) Donk) входит в число редких видов макромицетов, имеющих высокий природоохранный статус во многих странах Европы. Этот гриб развивается на старовозрастных дубах (Quercus spp.), и его численность существенно сокращается по мере уничтожения коренных дубрав [2; 5; 13; 15-19; 21; 22]. В Украине вид Piptoporus quercinus известен всего по нескольким находкам, и до недавнего времени был обнаружен на территории двух ботанико-географических регионов страны: Карпат и Закарпатья [1; 4]. В период 2002-2006 гг. этот вид был впервые обнаружен нами в Левобережной Украине: на валежных стволах и пнях Quercus robur L. в различных кварталах Национального природного парка «Гомольшанские леса». Учитывая редкость этого вида в общемировом масштабе, мы считаем необходимым привести подробную морфолого-таксономическую и экологическую характеристику этого вида, описать локалитеты его обнаружения на территории парка, а также мероприятия, необходимые для его сохранения. -

Diversity of Polyporales in the Malay Peninsular and the Application of Ganoderma Australe (Fr.) Pat
DIVERSITY OF POLYPORALES IN THE MALAY PENINSULAR AND THE APPLICATION OF GANODERMA AUSTRALE (FR.) PAT. IN BIOPULPING OF EMPTY FRUIT BUNCHES OF ELAEIS GUINEENSIS MOHAMAD HASNUL BIN BOLHASSAN FACULTY OF SCIENCE UNIVERSITY OF MALAYA KUALA LUMPUR 2013 DIVERSITY OF POLYPORALES IN THE MALAY PENINSULAR AND THE APPLICATION OF GANODERMA AUSTRALE (FR.) PAT. IN BIOPULPING OF EMPTY FRUIT BUNCHES OF ELAEIS GUINEENSIS MOHAMAD HASNUL BIN BOLHASSAN THESIS SUBMITTED IN FULFILMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY INSTITUTE OF BIOLOGICAL SCIENCES FACULTY OF SCIENCE UNIVERSITY OF MALAYA KUALA LUMPUR 2013 UNIVERSITI MALAYA ORIGINAL LITERARY WORK DECLARATION Name of Candidate: MOHAMAD HASNUL BIN BOLHASSAN (I.C No: 830416-13-5439) Registration/Matric No: SHC080030 Name of Degree: DOCTOR OF PHILOSOPHY Title of Project Paper/Research Report/Disertation/Thesis (“this Work”): DIVERSITY OF POLYPORALES IN THE MALAY PENINSULAR AND THE APPLICATION OF GANODERMA AUSTRALE (FR.) PAT. IN BIOPULPING OF EMPTY FRUIT BUNCHES OF ELAEIS GUINEENSIS. Field of Study: MUSHROOM DIVERSITY AND BIOTECHNOLOGY I do solemnly and sincerely declare that: 1) I am the sole author/writer of this work; 2) This Work is original; 3) Any use of any work in which copyright exists was done by way of fair dealing and for permitted purposes and any excerpt or extract from, or reference to or reproduction of any copyright work has been disclosed expressly and sufficiently and the title of the Work and its authorship have been acknowledge in this Work; 4) I do not have any actual -

From Taiwanofungus Camphoratus
Hindawi Publishing Corporation Evidence-Based Complementary and Alternative Medicine Volume 2011, Article ID 750230, 13 pages doi:10.1155/2011/750230 Research Article Apoptotic Cell Death and Inhibition of Wnt/β-Catenin Signaling Pathway in Human Colon Cancer Cells by an Active Fraction (HS7) from Taiwanofungus camphoratus Chi-Tai Yeh,1, 2, 3 Chih-Jung Yao,2, 4 Jiann-Long Yan,5 Shuang-En Chuang,5 Liang-Ming Lee,4 Chien-Ming Chen,1 Chuan-Feng Yeh,5 Chi-Han Li,5 and Gi-Ming Lai1, 2, 4, 5 1 Cancer Center, Shuang Ho Hospital, Taipei Medical University, Taipei 235, Taiwan 2 Center of Excellence for Cancer Research, Taipei Medical University, Taipei 110, Taiwan 3 Graduate Institute of Clinical Medicine, Taipei Medical University, Taipei 110, Taiwan 4 Cancer Center, Wan Fang Hospital, Taipei Medical University, Taipei 116, Taiwan 5 National Institute of Cancer Research, National Health Research Institutes, Miaoli 350, Taiwan Correspondence should be addressed to Gi-Ming Lai, [email protected] Received 27 September 2010; Revised 6 January 2011; Accepted 11 January 2011 Copyright © 2011 Chi-Tai Yeh et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Aberrant activation of Wnt/β-catenin signaling plays an important role in the development of colon cancer. HS7 is an active fraction extracted from Taiwanofungus camphoratus, which had been widely used as complementary medicine for Taiwan cancer patients in the past decades. In this study, we demonstrated the effects of HS7 on the growth inhibition, apoptosis induction, and Wnt/β-catenin signaling suppression in human colon cancer cells. -

Phylogenetic Classification of Trametes
TAXON 60 (6) • December 2011: 1567–1583 Justo & Hibbett • Phylogenetic classification of Trametes SYSTEMATICS AND PHYLOGENY Phylogenetic classification of Trametes (Basidiomycota, Polyporales) based on a five-marker dataset Alfredo Justo & David S. Hibbett Clark University, Biology Department, 950 Main St., Worcester, Massachusetts 01610, U.S.A. Author for correspondence: Alfredo Justo, [email protected] Abstract: The phylogeny of Trametes and related genera was studied using molecular data from ribosomal markers (nLSU, ITS) and protein-coding genes (RPB1, RPB2, TEF1-alpha) and consequences for the taxonomy and nomenclature of this group were considered. Separate datasets with rDNA data only, single datasets for each of the protein-coding genes, and a combined five-marker dataset were analyzed. Molecular analyses recover a strongly supported trametoid clade that includes most of Trametes species (including the type T. suaveolens, the T. versicolor group, and mainly tropical species such as T. maxima and T. cubensis) together with species of Lenzites and Pycnoporus and Coriolopsis polyzona. Our data confirm the positions of Trametes cervina (= Trametopsis cervina) in the phlebioid clade and of Trametes trogii (= Coriolopsis trogii) outside the trametoid clade, closely related to Coriolopsis gallica. The genus Coriolopsis, as currently defined, is polyphyletic, with the type species as part of the trametoid clade and at least two additional lineages occurring in the core polyporoid clade. In view of these results the use of a single generic name (Trametes) for the trametoid clade is considered to be the best taxonomic and nomenclatural option as the morphological concept of Trametes would remain almost unchanged, few new nomenclatural combinations would be necessary, and the classification of additional species (i.e., not yet described and/or sampled for mo- lecular data) in Trametes based on morphological characters alone will still be possible. -

A New Species of Antrodia (Basidiomycota, Polypores) from China
Mycosphere 8(7): 878–885 (2017) www.mycosphere.org ISSN 2077 7019 Article Doi 10.5943/mycosphere/8/7/4 Copyright © Guizhou Academy of Agricultural Sciences A new species of Antrodia (Basidiomycota, Polypores) from China Chen YY, Wu F* Institute of Microbiology, Beijing Forestry University, Beijing 100083, China Chen YY, Wu F 2017 –A new species of Antrodia (Basidiomycota, Polypores) from China. Mycosphere 8(7), 878–885, Doi 10.5943/mycosphere/8/7/4 Abstract A new species, Antrodia monomitica sp. nov., is described and illustrated from China based on morphological characters and molecular evidence. It is characterized by producing annual, fragile and nodulose basidiomata, a monomitic hyphal system with clamp connections on generative hyphae, hyaline, thin-walled and fusiform to mango-shaped basidiospores (6–7.5 × 2.3– 3 µm), and causing a typical brown rot. In phylogenetic analysis inferred from ITS and nLSU rDNA sequences, the new species forms a distinct lineage in the Antrodia s. l., and has a close relationship with A. oleracea. Key words – Fomitopsidaceae – phylogenetic analysis – taxonomy – wood-decaying fungi Introduction Antrodia P. Karst., typified with Polyporus serpens Fr. (=Antrodia albida (Fr.) Donk (Donk 1960, Ryvarden 1991), is characterized by a resupinate to effused-reflexed growth habit, white or pale colour of the context, a dimitic hyphal system with clamp connections on generative hyphae, hyaline, thin-walled, cylindrical to very narrow ellipsoid basidiospores which are negative in Melzer’s reagent and Cotton Blue, and causing a brown rot (Ryvarden & Melo 2014). Antrodia is a highly heterogeneous genus which is closely related to Fomitopsis P. -

A Phylogenetic Overview of the Antrodia Clade (Basidiomycota, Polyporales)
Mycologia, 105(6), 2013, pp. 1391–1411. DOI: 10.3852/13-051 # 2013 by The Mycological Society of America, Lawrence, KS 66044-8897 A phylogenetic overview of the antrodia clade (Basidiomycota, Polyporales) Beatriz Ortiz-Santana1 phylogenetic studies also have recognized the genera Daniel L. Lindner Amylocystis, Dacryobolus, Melanoporia, Pycnoporellus, US Forest Service, Northern Research Station, Center for Sarcoporia and Wolfiporia as part of the antrodia clade Forest Mycology Research, One Gifford Pinchot Drive, (SY Kim and Jung 2000, 2001; Binder and Hibbett Madison, Wisconsin 53726 2002; Hibbett and Binder 2002; SY Kim et al. 2003; Otto Miettinen Binder et al. 2005), while the genera Antrodia, Botanical Museum, University of Helsinki, PO Box 7, Daedalea, Fomitopsis, Laetiporus and Sparassis have 00014, Helsinki, Finland received attention in regard to species delimitation (SY Kim et al. 2001, 2003; KM Kim et al. 2005, 2007; Alfredo Justo Desjardin et al. 2004; Wang et al. 2004; Wu et al. 2004; David S. Hibbett Dai et al. 2006; Blanco-Dios et al. 2006; Chiu 2007; Clark University, Biology Department, 950 Main Street, Worcester, Massachusetts 01610 Lindner and Banik 2008; Yu et al. 2010; Banik et al. 2010, 2012; Garcia-Sandoval et al. 2011; Lindner et al. 2011; Rajchenberg et al. 2011; Zhou and Wei 2012; Abstract: Phylogenetic relationships among mem- Bernicchia et al. 2012; Spirin et al. 2012, 2013). These bers of the antrodia clade were investigated with studies also established that some of the genera are molecular data from two nuclear ribosomal DNA not monophyletic and several modifications have regions, LSU and ITS. A total of 123 species been proposed: the segregation of Antrodia s.l. -

Mating-Type Locus Organization and Mating-Type Chromosome Differentiation in the Bipolar Edible Button Mushroom Agaricus Bisporus
G C A T T A C G G C A T genes Article Mating-Type Locus Organization and Mating-Type Chromosome Differentiation in the Bipolar Edible Button Mushroom Agaricus bisporus Marie Foulongne-Oriol 1 , Ozgur Taskent 2, Ursula Kües 3, Anton S. M. Sonnenberg 4, Arend F. van Peer 4 and Tatiana Giraud 2,* 1 INRAE, MycSA, Mycologie et Sécurité des Aliments, 33882 Villenave d’Ornon, France; [email protected] 2 Ecologie Systématique Evolution, Bâtiment 360, CNRS, AgroParisTech, Université Paris-Saclay, 91400 Orsay, France; [email protected] 3 Molecular Wood Biotechnology and Technical Mycology, Goettingen Center for Molecular Biosciences (GZMB), Büsgen-Institute, University of Goettingen, Büsgenweg 2, 37077 Goettingen, Germany; [email protected] 4 Plant Breeding, Wageningen University and Research, Droevendaalsesteeg 1, 6708 PB Wageningen, The Netherlands; [email protected] (A.S.M.S.); [email protected] (A.F.v.P.) * Correspondence: [email protected] Abstract: In heterothallic basidiomycete fungi, sexual compatibility is restricted by mating types, typically controlled by two loci: PR, encoding pheromone precursors and pheromone receptors, and HD, encoding two types of homeodomain transcription factors. We analysed the single mating-type locus of the commercial button mushroom variety, Agaricus bisporus var. bisporus, and of the related Citation: Foulongne-Oriol, M.; variety burnettii. We identified the location of the mating-type locus using genetic map and genome Taskent, O.; Kües, U.; Sonnenberg, information, corresponding to the HD locus, the PR locus having lost its mating-type role. We A.S.M.; van Peer, A.F.; Giraud, T. -

Biodiversity of Wood-Decay Fungi in Italy
AperTO - Archivio Istituzionale Open Access dell'Università di Torino Biodiversity of wood-decay fungi in Italy This is the author's manuscript Original Citation: Availability: This version is available http://hdl.handle.net/2318/88396 since 2016-10-06T16:54:39Z Published version: DOI:10.1080/11263504.2011.633114 Terms of use: Open Access Anyone can freely access the full text of works made available as "Open Access". Works made available under a Creative Commons license can be used according to the terms and conditions of said license. Use of all other works requires consent of the right holder (author or publisher) if not exempted from copyright protection by the applicable law. (Article begins on next page) 28 September 2021 This is the author's final version of the contribution published as: A. Saitta; A. Bernicchia; S.P. Gorjón; E. Altobelli; V.M. Granito; C. Losi; D. Lunghini; O. Maggi; G. Medardi; F. Padovan; L. Pecoraro; A. Vizzini; A.M. Persiani. Biodiversity of wood-decay fungi in Italy. PLANT BIOSYSTEMS. 145(4) pp: 958-968. DOI: 10.1080/11263504.2011.633114 The publisher's version is available at: http://www.tandfonline.com/doi/abs/10.1080/11263504.2011.633114 When citing, please refer to the published version. Link to this full text: http://hdl.handle.net/2318/88396 This full text was downloaded from iris - AperTO: https://iris.unito.it/ iris - AperTO University of Turin’s Institutional Research Information System and Open Access Institutional Repository Biodiversity of wood-decay fungi in Italy A. Saitta , A. Bernicchia , S. P. Gorjón , E. -

Fungal Diversity in the Mediterranean Area
Fungal Diversity in the Mediterranean Area • Giuseppe Venturella Fungal Diversity in the Mediterranean Area Edited by Giuseppe Venturella Printed Edition of the Special Issue Published in Diversity www.mdpi.com/journal/diversity Fungal Diversity in the Mediterranean Area Fungal Diversity in the Mediterranean Area Editor Giuseppe Venturella MDPI • Basel • Beijing • Wuhan • Barcelona • Belgrade • Manchester • Tokyo • Cluj • Tianjin Editor Giuseppe Venturella University of Palermo Italy Editorial Office MDPI St. Alban-Anlage 66 4052 Basel, Switzerland This is a reprint of articles from the Special Issue published online in the open access journal Diversity (ISSN 1424-2818) (available at: https://www.mdpi.com/journal/diversity/special issues/ fungal diversity). For citation purposes, cite each article independently as indicated on the article page online and as indicated below: LastName, A.A.; LastName, B.B.; LastName, C.C. Article Title. Journal Name Year, Article Number, Page Range. ISBN 978-3-03936-978-2 (Hbk) ISBN 978-3-03936-979-9 (PDF) c 2020 by the authors. Articles in this book are Open Access and distributed under the Creative Commons Attribution (CC BY) license, which allows users to download, copy and build upon published articles, as long as the author and publisher are properly credited, which ensures maximum dissemination and a wider impact of our publications. The book as a whole is distributed by MDPI under the terms and conditions of the Creative Commons license CC BY-NC-ND. Contents About the Editor .............................................. vii Giuseppe Venturella Fungal Diversity in the Mediterranean Area Reprinted from: Diversity 2020, 12, 253, doi:10.3390/d12060253 .................... 1 Elias Polemis, Vassiliki Fryssouli, Vassileios Daskalopoulos and Georgios I.