Молекулярная Филогения Hamamelidaceae S.L. На Основе
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

P. Ramorum Chronology
A CHRONOLOGY OF PHYTOPHTHORA RAMORUM, CAUSE OF SUDDEN OAK DEATH AND OTHER FOLIAR DISEASES 8/21 • Oregon crews have conducted intensive ground surveys in the Port Orford area to determine the extent of the NA2 lineage infestation and found that it is larger than initially realized. Since late May, over 136 positive P. ramorum samples have been collected from tanoaks and rhododendrons surrounding the initial two tanoak trees along Highway 101. A 600-ft treatment buffer has been set around all infected trees resulting in a 475-acre treatment area. The Oregon Legislature has allocated $1.7 million for detection and treatment of sudden oak death over the next two years with a further $190,000 coming in 2021 from the U.S. Forest Service under a cooperative agreement. Treating this area is estimated to cost about $1.7 million. • USDA APHIS and APHIS-accredited laboratories have confirmed 128 positive samples for P. ramorum in various establishments (regulatory incidents) thus far in 2021. Rhododendron plants continue to be the most commonly detected infected genus, comprising 61.7% of the positive samples confirmed so far this year. The next most common genera to test positive were Pieris and Loropetalum, with 13.3% and 9.4% respectively. • In WA this May, regulatory staff performed a 2-day certification survey at a wholesale shipping nursery under compliance; 413 samples were collected and all samples tested negative for P. ramorum. In June, WSDA conducted a trace-forward investigation on plants shipped from a positive out-of-state nursery. Eight receiving locations in Washington were inspected. -

National List of Vascular Plant Species That Occur in Wetlands 1996
National List of Vascular Plant Species that Occur in Wetlands: 1996 National Summary Indicator by Region and Subregion Scientific Name/ North North Central South Inter- National Subregion Northeast Southeast Central Plains Plains Plains Southwest mountain Northwest California Alaska Caribbean Hawaii Indicator Range Abies amabilis (Dougl. ex Loud.) Dougl. ex Forbes FACU FACU UPL UPL,FACU Abies balsamea (L.) P. Mill. FAC FACW FAC,FACW Abies concolor (Gord. & Glend.) Lindl. ex Hildebr. NI NI NI NI NI UPL UPL Abies fraseri (Pursh) Poir. FACU FACU FACU Abies grandis (Dougl. ex D. Don) Lindl. FACU-* NI FACU-* Abies lasiocarpa (Hook.) Nutt. NI NI FACU+ FACU- FACU FAC UPL UPL,FAC Abies magnifica A. Murr. NI UPL NI FACU UPL,FACU Abildgaardia ovata (Burm. f.) Kral FACW+ FAC+ FAC+,FACW+ Abutilon theophrasti Medik. UPL FACU- FACU- UPL UPL UPL UPL UPL NI NI UPL,FACU- Acacia choriophylla Benth. FAC* FAC* Acacia farnesiana (L.) Willd. FACU NI NI* NI NI FACU Acacia greggii Gray UPL UPL FACU FACU UPL,FACU Acacia macracantha Humb. & Bonpl. ex Willd. NI FAC FAC Acacia minuta ssp. minuta (M.E. Jones) Beauchamp FACU FACU Acaena exigua Gray OBL OBL Acalypha bisetosa Bertol. ex Spreng. FACW FACW Acalypha virginica L. FACU- FACU- FAC- FACU- FACU- FACU* FACU-,FAC- Acalypha virginica var. rhomboidea (Raf.) Cooperrider FACU- FAC- FACU FACU- FACU- FACU* FACU-,FAC- Acanthocereus tetragonus (L.) Humm. FAC* NI NI FAC* Acanthomintha ilicifolia (Gray) Gray FAC* FAC* Acanthus ebracteatus Vahl OBL OBL Acer circinatum Pursh FAC- FAC NI FAC-,FAC Acer glabrum Torr. FAC FAC FAC FACU FACU* FAC FACU FACU*,FAC Acer grandidentatum Nutt. -

Integrating Palaeontological and Molecular Data Uncovers Multiple
Integrating palaeontological and molecular data uncovers multiple ancient and recent dispersals in the pantropical Hamamelidaceae Xiaoguo Xiang, Kunli Xiang, Rosa del C. Ortiz, Florian Jabbour, Wei Wang To cite this version: Xiaoguo Xiang, Kunli Xiang, Rosa del C. Ortiz, Florian Jabbour, Wei Wang. Integrating palaeontolog- ical and molecular data uncovers multiple ancient and recent dispersals in the pantropical Hamamel- idaceae. Journal of Biogeography, Wiley, 2019, 46 (11), pp.2622-2631. 10.1111/jbi.13690. hal- 02612865 HAL Id: hal-02612865 https://hal.archives-ouvertes.fr/hal-02612865 Submitted on 19 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Integrating palaeontological and molecular data uncovers multiple ancient and recent dispersals in the pantropical Hamamelidaceae Xiaoguo Xiang1,2, Kunli Xiang1,3, Rosa Del C. Ortiz4, Florian Jabbour5, Wei Wang1,3 1State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing, China 2Jiangxi Province Key Laboratory of Watershed Ecosystem -

The Vascular Flora of Sandy Run Savannas State Natural Area, Onslow and Pender Counties, North Carolina --In Press-- John B
The Vascular Flora of Sandy Run Savannas State Natural Area, Onslow and Pender Counties, North Carolina --In Press-- John B. Taggart Department of Environmental Studies, University of North Carolina at Wilmington, 601 South College Road, Wilmington, North Carolina 28403 ______________________________________________________________________________ ABSTRACT The vascular plants of Sandy Run Savannas State Natural Area, located in portions of Onslow and Pender counties, North Carolina, are presented as an annotated species list. A total of 590 taxa in 315 genera and 119 families were collected from eight plant communities. Families with the highest numbers of species were the Asteraceae (80), Poaceae (66), and Cyperaceae (65). Two species, Carex lutea (golden sedge) and Thalictrum cooleyi (Cooley’s meadowrue), have federal endangered status. A total of 23 taxa are tracked by the North Carolina Natural Heritage Program, while 29 others are considered rare, but not included on the priority list. Of 44 species considered strict endemic or near-endemic taxa to the North and South Carolina Coastal Plain, 18 (41%) were collected in this study. Selected pine savannas within the site were rated as nationally significant by the North Carolina Natural Heritage Program. Fifty-one (51) non-native species were present and represented 8.7 % of the flora. _________________________________________________________________________ INTRODUCTION Sandy Run Savannas State Natural Area encompasses portions of western Onslow and northeastern Pender counties in North Carolina. State acquisition of this coastal plain site began in 2007 as a cooperative effort between The Nature Conservancy in North Carolina and the North Carolina Division of Parks and Recreation to protect approximately 1,214 ha comprised of seven tracts (Figure 1). -

Clarifying Taxonomy and Nomenclature of Fothergilla
HORTSCIENCE 42(3):470–473. 2007. wide and from 0.9 to 1.5 mm deep. Cytology determined a chromosome number of 2n = 4x = 48 (Weaver, Jr., 1969). In contrast, F. Clarifying Taxonomy and major is found on upland sites in the pied- mont and mountains of North Carolina, South Nomenclature of Fothergilla Carolina, Georgia, Alabama, Tennessee, and Arkansas (Flora of North America Editorial (Hamamelidaceae) Cultivars Committee, 1993+; Weakley, 2006; Weaver, Jr., 1969). This species generally is larger in stature (7–65 dm) than F. gardenii and is and Hybrids distinguished by larger leaves ranging from Thomas G. Ranney1,4 and Nathan P. Lynch2 2.5 to 13 cm long and 4.2 to 12.5 cm wide Department of Horticultural Science, Mountain Horticultural Crops that generally are toothed from below the middle and conspicuously asymmetric at Research and Extension Center, North Carolina State University, the base. Stipules are 2.8–7 (10.2) mm 455 Research Drive, Fletcher, NC 28732-9244 long. Stamens generally number (18) 22–32. 1 The hypanthium at anthesis ranges from Paul R. Fantz 2.4 to 3.9 mm wide and from 1.5 to 3 mm Department of Horticultural Science, North Carolina State University, deep. Cytology determined a chromosome Raleigh, NC 27695-7609 number of 2n =6x = 72 (Weaver, Jr., 1969). Although there is no known diploid species 3 Paul Cappiello of fothergilla, Parrotiopsis (Niedenzu) Yew Dell Gardens, P.O. Box 1334, 6220 Old LaGrange Road, Crestwood, C. Schneid. is a closely allied genus with KY 40014-9550 2n =2x = 24 (Goldblatt and Endress, 1977; Li and Bogle, 2001; Weaver, Jr., 1969) and Additional index words. -
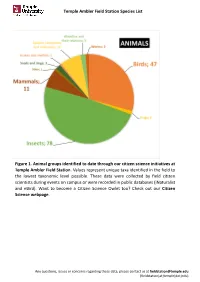
Temple Ambler Field Station Species List Figure 1. Animal Groups Identified to Date Through Our Citizen Science Initiatives at T
Temple Ambler Field Station Species List Figure 1. Animal groups identified to date through our citizen science initiatives at Temple Ambler Field Station. Values represent unique taxa identified in the field to the lowest taxonomic level possible. These data were collected by field citizen scientists during events on campus or were recorded in public databases (iNaturalist and eBird). Want to become a Citizen Science Owlet too? Check out our Citizen Science webpage. Any questions, issues or concerns regarding these data, please contact us at [email protected] (fieldstation[at}temple[dot]edu) Temple Ambler Field Station Species List Figure 2. Plant diversity identified to date in the natural environments and designed gardens of the Temple Ambler Field Station and Ambler Arboretum. These values represent unique taxa identified to the lowest taxonomic level possible. Highlighted are 14 of the 116 flowering plant families present that include 524 taxonomic groups. A full list can be found in our species database. Cultivated specimens in our Greenhouse were not included here. Any questions, issues or concerns regarding these data, please contact us at [email protected] (fieldstation[at}temple[dot]edu) Temple Ambler Field Station Species List database_title Temple Ambler Field Station Species List last_update 22October2020 description This database includes all species identified to their lowest taxonomic level possible in the natural environments and designed gardens on the Temple Ambler campus. These are occurrence records and each taxon is only entered once. This is an occurrence record, not an abundance record. IDs were performed by senior scientists and specialists, as well as citizen scientists visiting campus. -

Fothergilla Gardenii1
Fact Sheet FPS-214 October, 1999 Fothergilla gardenii1 Edward F. Gilman2 Introduction This 4- to 6-feet tall shrub covers itself with soft, white flowers each spring before leaves emerge (Fig. 1). It appears to be covered with snow when in full bloom. Bright red, orange or yellow fall color bring the shrub back to life before leaves fall to the ground. General Information Scientific name: Fothergilla gardenii Pronunciation: faw-thur-GIL-luh gar-DEE-nee-eye Common name(s): Dwarf Fothergilla Family: Hamamelidaceae Plant type: shrub USDA hardiness zones: 5 through 8A (Fig. 2) Planting month for zone 7: year round Planting month for zone 8: year round Planting month for zone 9: year round Planting month for zone 10 and 11: year round Origin: native to North America Uses: accent; border; mass planting Figure 1. Dwarf Fothergilla. Availablity: somewhat available, may have to go out of the region to find the plant Texture: medium Description Foliage Height: 4 to 6 feet Spread: 4 to 6 feet Leaf arrangement: alternate Plant habit: round Leaf type: simple Plant density: dense Leaf margin: dentate Growth rate: slow Leaf shape: elliptic (oval); obovate; orbiculate 1.This document is Fact Sheet FPS-214, one of a series of the Environmental Horticulture Department, Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida. Publication date: October, 1999 Please visit the EDIS Web site at http://edis.ifas.ufl.edu. 2. Edward F. Gilman, professor, Environmental Horticulture Department, Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida, Gainesville, 32611. -

Functional Integration of Floral Plant Traits: Shape and Symmetry, Optical Signal, Reward and Reproduction in the Angiosperm Flower
Functional Integration of Floral Plant Traits: Shape and Symmetry, Optical Signal, Reward and Reproduction in the Angiosperm Flower Dissertation zur Erlangung des Doktorgrades (Dr. rer. nat.) der Mathematisch-Naturwissenschaftlichen Fakultät der Rheinischen Friedrich-Wilhelms-Universität Bonn vorgelegt von Andreas Wilhelm Mues aus Kirchhellen Bonn, den 20. Januar 2020 1 2 Angefertigt mit Genehmigung der Mathematisch-Naturwissenschaftlichen Fakultät der Rheinischen Friedrich-Wilhelms-Universität Bonn Erstgutachter: Prof. Dr. Maximilian Weigend, Universität Bonn Zweitgutachter: Prof. Dr. Eberhard Fischer, Universität Koblenz Tag der Promotion: 30. April 2020 Erscheinungsjahr: 2020 3 4 Acknowledgements I thank Prof. Dr. Maximilian Weigend, supervisor, for his guidance and support, and for giving me the opportunity to study the holistic subject of floral functional integration and plant-animal interaction. I am grateful for the experience and for the research agendas he entrusted to me: Working with the extensive Living Collections of Bonn Botanical Gardens was an honour, and I have learned a lot. I thank Prof. Dr. Eberhard Fisher, for agreeing to be my second supervisor, his advice and our shared passion for the plant world. I would like to thank many people of the Nees Institute and Bonn Botanical Gardens who contributed to this work and who gave me good memories of my years of study: I thank Lisabeth Hoff, Tianjun Liu, Luisa Sophie Nicolin and Simon Brauwers for their contribution in collecting shares of the raw data together with me, and for being eager students – especially counting pollen and ovule numbers and measuring nectar reward was a test of patience sometimes, and we have counted and measured a lot … Thank you! Special thanks go to Gardeners of the Bonn Botanical Gardens, for their constant support throughout the years, their love for the plant world in general and their commitment and care for the Living Collection: Klaus Mahlberg (Streptocarpus), Birgit Emde (carnivorous plants), Klaus Bahr (Geraniales), Bernd Reinken and Klaus Michael Neumann. -

Adiciones Y Comentarios a La Flora De Honduras Cyril H. Nelson
Adiciones y Comentarios a la Flora de Honduras Cyril H. Nelson1 Resumen. Se agregan y comentan taxones de la flora de Honduras, de los cuales hay 10 familias nuevas, 52 géneros, 338 especies, nueve subespecies, cinco variedades, una forma, 19 endemismos, 53 sinonimias y 28 nombres comunes. Palabras clave: Familias, géneros, sinonimia, endemismo, nuevos registros, nombres comunes. Adiciones y Comentarios a la Flora de Honduras Abstract. Taxa of the flora of Honduras, of which there are 10 new families, 52 genera, 338 species, nine subspecies, five varieties, one form, 19 endemisms, 53 synonyms, and 28 common names are added and commented. Kew words: Families, genera, synonymy, endemism, new records, common names. Introducción el herbario del Jardín Botánico de Missouri (MO) es donde están depositadas la mayoría de las especies Desde la publicación de Nelson Sutherland citadas. Sin embargo, no necesariamente es el único (2008) del Catálogo de las Plantas Vasculares de herbario donde hay duplicados, pudiéndose encontrar Honduras han aparecido nuevos registros para la flora también en el herbario de la Escuela Agrícola de Honduras, ya sea por la publicación de especies Panamericana (EAP). La Flora Mesomericana nuevas, ya sea por la corrección de especímenes mal (Davidse et al., 1994-) aún no completada, es una identificados, ya sea por la identificación de antiguos referencia para saber la aceptación o no de un taxon. especímenes que reposaban en herbarios esperando que un especialista los identificara. También la Resultados publicación del grupo APG III (2009) ha contribuido al reordenamiento de órdenes y familias en las plantas A continuación se presenta un listado de con flores. -

EXCLUDED TAXA Digital Atlas of the Virginia Flora Current As of June 5, 2021 © Virginia Botanical Associates 2021
EXCLUDED TAXA Digital Atlas of the Virginia Flora Current as of June 5, 2021 © Virginia Botanical Associates 2021 INTRODUCTION The following is an annotated list of more than 300 taxa that have been reported for Virginia but are currently excluded from mapping in the Digital Atlas. The reasons for their exclusion fall into several categories, the most common of which are 1) lack of supporting voucher specimens, or 2) lack of evidence that a taxon meets the basic criteria for establishment described in About the Digital Atlas. Also included are species that are known or suspected to be errors based on misidentifications. VBA decided to post this list in order to call attention to these taxa, generate new information, and reduce the perpetuation of misinformation that has characterized many of the entries. Massey (1961) reported many species from Virginia without known documentation or based on errors of interpretation of the "Gray's manual range." Despite years of cleaning up these errors and unsubstantiated taxa in the first three editions of the Atlas, many of them were given a new lease on life by Kartesz (1999), and from there by other sources such as USDA Plants and iterative drafts of Weakley's regional flora (most of these errors have been corrected in the current (2015), online version of Kartesz's North American Plant Atlas). Previous errors mapped in hard-copy Atlas editions and the Digital Atlas have also been repeated by various sources. Today, modern technology and search engines can and will find the most obscure details from centuries of botanical literature, with no ability to distinguish valid from erroneous contexts. -

Systematics of the Hamamelidaceae Based on Morphological and Molecular Evidence Jianhua Li University of New Hampshire, Durham
University of New Hampshire University of New Hampshire Scholars' Repository Doctoral Dissertations Student Scholarship Winter 1997 Systematics of the Hamamelidaceae based on morphological and molecular evidence Jianhua Li University of New Hampshire, Durham Follow this and additional works at: https://scholars.unh.edu/dissertation Recommended Citation Li, Jianhua, "Systematics of the Hamamelidaceae based on morphological and molecular evidence" (1997). Doctoral Dissertations. 1997. https://scholars.unh.edu/dissertation/1997 This Dissertation is brought to you for free and open access by the Student Scholarship at University of New Hampshire Scholars' Repository. It has been accepted for inclusion in Doctoral Dissertations by an authorized administrator of University of New Hampshire Scholars' Repository. For more information, please contact [email protected]. f INFORMATION TO USERS This manuscript has been reproduced from the microfilm master. UMI films the text directly from the original or copy submitted. Thus, some thesis and dissertation copies are in typewriter face, while others may be from any type of computer printer. The quality of this reproduction is dependent upon the quality of the copy submitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleedthrough, substandard margins, and improper alignment can adversely affect reproduction. In the unlikely event that the author did not send UMI a complete manuscript and there are missing pages, these will be noted. Also, if unauthorized copyright material had to be removed, a note will indicate the deletion. Oversize materials (e.g., maps, drawings, charts) are reproduced by sectioning the original, beginning at the upper left-hand comer and continuing from left to right in equal sections with small overlaps. -
A New Paralog Removal Pipeline Resolves Conflict Between RAD-Seq and Enrichment
bioRxiv preprint doi: https://doi.org/10.1101/2020.10.26.355248; this version posted October 27, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 A New Paralog Removal Pipeline Resolves Conflict between RAD-seq and Enrichment 2 Wenbin Zhou1*, John Soghigian2, Qiu-yun (Jenny) Xiang1* 3 1 Department of Plant and Microbial Biology, North Carolina State University, Raleigh, NC 4 27965, USA 5 2 Department of Entomology and Plant Pathology, North Carolina State University, Raleigh, NC 6 27965, USA 7 8 *Corresponding authors e-mail addresses: [email protected]; [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2020.10.26.355248; this version posted October 27, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. ZHOU ET AL. 9 ABSTRACT 10 Target enrichment and RAD-seq are well-established high throughput sequencing 11 technologies that have been increasingly used for phylogenomic studies, and the choice between 12 methods is a practical issue for plant systematists studying the evolutionary histories of 13 biodiversity of relatively recent origins. However, few studies have compared the congruence and 14 conflict between results from the two methods within the same group of organisms, especially in 15 plants, where extensive genome duplication events may complicate phylogenomic 16 analyses.