Starvation Following Cobalt-60 Γ-Ray Radiation Enhances Metastasis in U251 Glioma Cells by Regulating the Transcription Factor SP1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Development and Maintenance of Epidermal Stem Cells in Skin Adnexa
International Journal of Molecular Sciences Review Development and Maintenance of Epidermal Stem Cells in Skin Adnexa Jaroslav Mokry * and Rishikaysh Pisal Medical Faculty, Charles University, 500 03 Hradec Kralove, Czech Republic; [email protected] * Correspondence: [email protected] Received: 30 October 2020; Accepted: 18 December 2020; Published: 20 December 2020 Abstract: The skin surface is modified by numerous appendages. These structures arise from epithelial stem cells (SCs) through the induction of epidermal placodes as a result of local signalling interplay with mesenchymal cells based on the Wnt–(Dkk4)–Eda–Shh cascade. Slight modifications of the cascade, with the participation of antagonistic signalling, decide whether multipotent epidermal SCs develop in interfollicular epidermis, scales, hair/feather follicles, nails or skin glands. This review describes the roles of epidermal SCs in the development of skin adnexa and interfollicular epidermis, as well as their maintenance. Each skin structure arises from distinct pools of epidermal SCs that are harboured in specific but different niches that control SC behaviour. Such relationships explain differences in marker and gene expression patterns between particular SC subsets. The activity of well-compartmentalized epidermal SCs is orchestrated with that of other skin cells not only along the hair cycle but also in the course of skin regeneration following injury. This review highlights several membrane markers, cytoplasmic proteins and transcription factors associated with epidermal SCs. Keywords: stem cell; epidermal placode; skin adnexa; signalling; hair pigmentation; markers; keratins 1. Epidermal Stem Cells as Units of Development 1.1. Development of the Epidermis and Placode Formation The embryonic skin at very early stages of development is covered by a surface ectoderm that is a precursor to the epidermis and its multiple derivatives. -

Peking University-Juntendo University Joint Symposium on Cancer Research and Treatment ADAM28 (A Disintegrin and Metalloproteinase 28) in Cancer Cell Proliferation and Progression
Whatʼs New from Juntendo University, Tokyo Juntendo Medical Journal 2017. 63(5), 322-325 Peking University - Juntendo University Joint Symposium on Cancer Research and Treatment ADAM28 (a Disintegrin and Metalloproteinase 28) in Cancer Cell Proliferation and Progression YASUNORI OKADA* *Department of Pathophysiology for Locomotive and Neoplastic Diseases, Juntendo University Graduate School of Medicine, Tokyo, Japan A disintegrinandmetalloproteinase 28 (ADAM28) is overexpressedpredominantlyby carcinoma cells in more than 70% of the non-small cell lung carcinomas, showing positive correlations with carcinoma cell proliferation and metastasis. ADAM28 cleaves insulin-like growth factor binding protein-3 (IGFBP-3) in the IGF-I/IGFBP-3 complex, leading to stimulation of cell proliferation by intact IGF-I released from the complex. ADAM28 also degrades von Willebrand factor (VWF), which induces apoptosis in human carcinoma cell lines with negligible ADAM28 expression, andthe VWF digestionby ADAM28-expressing carcinoma cells facilitates them to escape from VWF-induced apoptosis, resulting in promotion of metastasis. We have developed human antibodies against ADAM28 andshown that one of them significantly inhibits tumor growth andmetastasis using lung adenocarcinoma cells. Our data suggest that ADAM28 may be a new molecular target for therapy of the patients with ADAM28-expressing non-small cell lung carcinoma. Key words: a disintegrin and metalloproteinase 28 (ADAM28), cell proliferation, invasion, metastasis, human antibody inhibitor Introduction human cancers 2). However, development of the synthetic inhibitors of MMPs andtheir application Cancer cell proliferation andprogression are for treatment of the cancer patients failed 3). modulated by proteolytic cleavage of tissue micro- On the other hand, members of the ADAM (a environmental factors such as extracellular matrix disintegrin and metalloproteinase) gene family, (ECM), growth factors andcytokines, receptors another family belonging to the metzincin gene andcell adhesionmolecules. -

Innate Pro–B-Cell Progenitors Protect Against Type 1 Diabetes By
Innate pro–B-cell progenitors protect against type 1 PNAS PLUS diabetes by regulating autoimmune effector T cells Ruddy Montandona,b,1, Sarantis Korniotisa,b,1, Esther Layseca-Espinosaa,b,2, Christophe Grasa,b, Jérôme Mégretc, Sophie Ezinea,d, Michel Dya,b, and Flora Zavalaa,b,3 aFaculté de Médecine Site Necker, Université Paris Descartes, bCentre National de la Recherche Scientifique Unité Mixte de Recherche 8147, 75015 Paris, France; cInstitut Fédératif de Recherche 94 Necker-Enfants Malades, 75015 Paris, France; and dInstitut National de la Santé et de la Recherche Médicale U1020, 75015 Paris, France Edited by Simon Fillatreau, Deutsches Rheuma-Forschungszentrum, Berlin, Germany, and accepted by the Editorial Board May 6, 2013 (received for review December 24, 2012) Diverse hematopoietic progenitors, including myeloid populations emergence of regulatory B cells (Bregs), along with acquired-type arising in inflammatory and tumoral conditions and multipotent stimulation, such as B-cell receptor (BCR) engagement concomi- cells, mobilized by hematopoietic growth factors or emerging during tant or not with CD40 activation (10, 11). Such induced regulatory parasitic infections, display tolerogenic properties. Innate immune B-cell functions are believed to be more robust than those ex- stimuli confer regulatory functions to various mature B-cell subsets pressed by naive and resting B cells, which can nevertheless tolerize but immature B-cell progenitors endowed with suppressive proper- naive T cells and induce regulatory T cells (Tregs) (12, 13). ties per se or after differentiating into more mature regulatory Bregs are a heterogeneous lymphocyte subset present among all B cells remain to be characterized. Herein we provide evidence for major B-cell populations (14–17). -

Human ADAM12 Quantikine ELISA
Quantikine® ELISA Human ADAM12 Immunoassay Catalog Number DAD120 For the quantitative determination of A Disintegrin And Metalloproteinase domain- containing protein 12 (ADAM12) concentrations in cell culture supernates, serum, plasma, and urine. This package insert must be read in its entirety before using this product. For research use only. Not for use in diagnostic procedures. TABLE OF CONTENTS SECTION PAGE INTRODUCTION .....................................................................................................................................................................1 PRINCIPLE OF THE ASSAY ...................................................................................................................................................2 LIMITATIONS OF THE PROCEDURE .................................................................................................................................2 TECHNICAL HINTS .................................................................................................................................................................2 MATERIALS PROVIDED & STORAGE CONDITIONS ...................................................................................................3 OTHER SUPPLIES REQUIRED .............................................................................................................................................3 PRECAUTIONS .........................................................................................................................................................................4 -

2021 Undergraduate Research Symposium Program
FORDHAM COLLEGE AT ROSE HILL 14TH ANNUAL UNDERGRADUATE RESEARCH SYMPOSIUM Wednesday, May 5, 2021 AN INTERDISCIPLINARY CELEBRATION OF OUR STUDENTS AND MENTORS The Fourteenth Annual Fordham College at Rose Hill Undergraduate Research Symposium Program | Spring 2021 Welcome to the Fourteenth Annual FCRH Research Symposium, for the first time in a hybrid format! The accomplishments of our students and mentors during the pandemic have been extraordinary and we are overjoyed to celebrate them today. From our beautiful campus, to their homes throughout the country and beyond, our undergraduate research community was always open for new discoveries. We are delighted to share 120 abstracts from over 200 students who pursued their projects during such challenging times. Their work, dedication, and determination to be a part of today’s event is inspiring and what FCRH undergraduate research is all about. We are in this together to urge each other on and findings shared today may well change the world. We are also proud to announce that the 11th volume of the Fordham Undergraduate Research Journal has been published. The FURJ team took on an enormous undertaking of running their operation in hybrid format, with a record number of submissions, and as always, they have dazzled us with the quality of their efforts. Undergraduate research has become a part of who we are at FCRH. Because of this, our program, against all odds in the past year, continues to grow, expanding across disciplines and accessible to all students in a number of ways. Students are creating new knowledge in our labs, independently with the guidance of their mentors, as part of innovative class projects, and even to support their activism. -

ADAM10 Site-Dependent Biology: Keeping Control of a Pervasive Protease
International Journal of Molecular Sciences Review ADAM10 Site-Dependent Biology: Keeping Control of a Pervasive Protease Francesca Tosetti 1,* , Massimo Alessio 2, Alessandro Poggi 1,† and Maria Raffaella Zocchi 3,† 1 Molecular Oncology and Angiogenesis Unit, IRCCS Ospedale Policlinico S. Martino Largo R. Benzi 10, 16132 Genoa, Italy; [email protected] 2 Proteome Biochemistry, IRCCS San Raffaele Scientific Institute, 20132 Milan, Italy; [email protected] 3 Division of Immunology, Transplants and Infectious Diseases, IRCCS San Raffaele Scientific Institute, 20132 Milan, Italy; [email protected] * Correspondence: [email protected] † These authors contributed equally to this work as last author. Abstract: Enzymes, once considered static molecular machines acting in defined spatial patterns and sites of action, move to different intra- and extracellular locations, changing their function. This topological regulation revealed a close cross-talk between proteases and signaling events involving post-translational modifications, membrane tyrosine kinase receptors and G-protein coupled recep- tors, motor proteins shuttling cargos in intracellular vesicles, and small-molecule messengers. Here, we highlight recent advances in our knowledge of regulation and function of A Disintegrin And Metalloproteinase (ADAM) endopeptidases at specific subcellular sites, or in multimolecular com- plexes, with a special focus on ADAM10, and tumor necrosis factor-α convertase (TACE/ADAM17), since these two enzymes belong to the same family, share selected substrates and bioactivity. We will discuss some examples of ADAM10 activity modulated by changing partners and subcellular compartmentalization, with the underlying hypothesis that restraining protease activity by spatial Citation: Tosetti, F.; Alessio, M.; segregation is a complex and powerful regulatory tool. -

BD Pharmingen™ PE Rat Anti-Human Integrin Β7
BD Pharmingen™ Technical Data Sheet PE Rat Anti-Human Integrin β7 Product Information Material Number: 555945 Size: 100 tests Vol. per Test: 20 µl Clone: FIB504 Isotype: Rat (SD) IgG2a, κ Reactivity: QC Testing: Human Workshop: VI A024, VI 6T-101 Storage Buffer: Aqueous buffered solution containing BSA and ≤0.09% sodium azide. Description FIB504 reacts with mouse integrin β7 subunit (130 kD) but also cross reacts with human integrin β7. Integrin β7 associates with α4 (CD49d) expressed on subsets of lymphocytes and thymus. It also associates with αIEL (CD103) expressed on T cells adjacent to mucosal epithelium and intraepithelial lymphocytes. Integrin β7 plays an important role in the adhesion of leukocytes to endothelial cells promoting the transmigration of leukocytes to extravascular spaces during the inflammatory response. Profile of peripheral blood lymphocytes analyzed on a FACScan (BDIS, San Jose, CA) Preparation and Storage The monoclonal antibody was purified from tissue culture supernatant or ascites by affinity chromatography. The antibody was conjugated with R-PE under optimum conditions, and unconjugated antibody and free PE were removed by gel filtration chromatography. Store undiluted at 4° C and protected from prolonged exposure to light. Do not freeze. Application Notes Application Flow cytometry Routinely Tested Suggested Companion Products Catalog Number Name Size Clone 555844 PE Rat IgG2a, κ Isotype Control 100 tests R35-95 555945 Rev. 3 Page 1 of 2 Product Notices 1. This reagent has been pre-diluted for use at the recommended Volume per Test. We typically use 1 X 10e6 cells in a 100-µl experimental sample (a test). 2. -

Smooth Muscle-Specific MMP17 (MT4-MMP) Defines the Intestinal ECM Niche 2 3 Mara Martín-Alonso1*, Håvard T
bioRxiv preprint doi: https://doi.org/10.1101/2020.06.18.147769; this version posted June 18, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Smooth muscle-specific MMP17 (MT4-MMP) defines the intestinal ECM niche 2 3 Mara Martín-Alonso1*, Håvard T. Lindholm1,# , Sharif Iqbal2,3,#, Pia Vornewald1#, Sigrid Hoel1, 4 Mirjam J. Damen4, A.F.Maarten Altelaar4, Pekka Katajisto2,3,5, Alicia G. Arroyo6,7, Menno J. 5 Oudhoff1* 6 7 1CEMIR – Centre of Molecular Inflammation Research, Department of Clinical and Molecular 8 Medicine, NTNU – Norwegian University of Science and Technology, 7491 Trondheim, 9 Norway 10 2Institute of Biotechnology, HiLIFE, University of Helsinki, Finland 11 3Molecular and Integrative Bioscience Research Programme, Faculty of Biological and 12 Environmental Sciences, University of Helsinki, Helsinki, Finland 13 4Biomolecular Mass Spectrometry and Proteomics, Utrecht University, Utrecht, The 14 Netherlands 15 5Department of Biosciences and Nutrition, Karolinska Institutet, Stockholm, Sweden 16 6Centro de Investigaciones Biológicas Margarita Salas (CIB-CSIC), Madrid, Spain 17 7Vascular Pathophysiology Area, Centro Nacional de Investigaciones Cardiovasculares 18 (CNIC), Madrid, Spain.CNIC, Madrid, Spain 19 20 #These authors contributed equally to this work 21 *corresponding authors, e-mail: [email protected], [email protected] 22 bioRxiv preprint doi: https://doi.org/10.1101/2020.06.18.147769; this version posted June 18, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. -

Molecular Pathways Associated with the Nutritional Programming of Plant-Based Diet Acceptance in Rainbow Trout Following an Early Feeding Exposure Mukundh N
Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure Mukundh N. Balasubramanian, Stéphane Panserat, Mathilde Dupont-Nivet, Edwige Quillet, Jérôme Montfort, Aurélie Le Cam, Françoise Médale, Sadasivam J. Kaushik, Inge Geurden To cite this version: Mukundh N. Balasubramanian, Stéphane Panserat, Mathilde Dupont-Nivet, Edwige Quillet, Jérôme Montfort, et al.. Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure. BMC Genomics, BioMed Central, 2016, 17 (1), pp.1-20. 10.1186/s12864-016-2804-1. hal-01346912 HAL Id: hal-01346912 https://hal.archives-ouvertes.fr/hal-01346912 Submitted on 19 Jul 2016 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Balasubramanian et al. BMC Genomics (2016) 17:449 DOI 10.1186/s12864-016-2804-1 RESEARCH ARTICLE Open Access Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure Mukundh N. Balasubramanian1, Stephane Panserat1, Mathilde Dupont-Nivet2, Edwige Quillet2, Jerome Montfort3, Aurelie Le Cam3, Francoise Medale1, Sadasivam J. Kaushik1 and Inge Geurden1* Abstract Background: The achievement of sustainable feeding practices in aquaculture by reducing the reliance on wild-captured fish, via replacement of fish-based feed with plant-based feed, is impeded by the poor growth response seen in fish fed high levels of plant ingredients. -
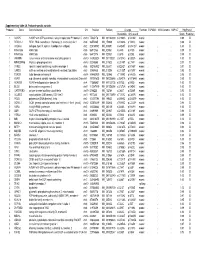
Supplementary Table 2A. Proband-Specific Variants Proband
Supplementary table 2A. Proband-specific variants Proband Gene Gene full name Chr Position RefSeqChange Function ESP6500 1000 Genome SNP137 PolyPhen2 Nucleotide Amino acid Score Prediction 1 AGAP5 ArfGAP with GTPase domain, ankyrin repeat and PH domain 5 chr10 75435178 NM_001144000 c.G1240A p.V414M exonic . 0.99 D 1 REXO1L1 REX1, RNA exonuclease 1 homolog (S. cerevisiae)-like 1 chr8 86574465 NM_172239 c.A1262G p.Y421C exonic . 0.98 D 1 COL4A3 collagen, type IV, alpha 3 (Goodpasture antigen) chr2 228169782 NM_000091 c.G4235T p.G1412V exonic . 1.00 D 1 KIAA1586 KIAA1586 chr6 56912133 NM_020931 c.C44G p.A15G exonic . 0.99 D 1 KIAA1586 KIAA1586 chr6 56912176 NM_020931 c.C87G p.D29E exonic . 0.99 D 1 JAKMIP3 Janus kinase and microtubule interacting protein 3 chr10 133955524 NM_001105521 c.G1574C p.G525A exonic . 1.00 D 1 MPHOSPH8 M-phase phosphoprotein 8 chr13 20240685 NM_017520 c.C2140T p.L714F exonic . 0.97 D 1 SYNE1 spectrin repeat containing, nuclear envelope 1 chr6 152783902 NM_033071 c.A2242T p.N748Y exonic . 0.97 D 1 CAND2 cullin-associated and neddylation-dissociated 2 (putative) chr3 12858835 NM_012298 c.C2125T p.H709Y exonic . 0.95 D 1 TDRD9 tudor domain containing 9 chr14 104460909 NM_153046 c.T1289G p.V430G exonic . 0.96 D 2 ASPM asp (abnormal spindle) homolog, microcephaly associated (Drosophila)chr1 197091625 NM_001206846 c.G3491A p.R1164H exonic . 0.99 D 2 ADAM29 ADAM metallopeptidase domain 29 chr4 175896951 NM_001130705 c.A275G p.D92G exonic . 1.00 D 2 BCO2 beta-carotene oxygenase 2 chr11 112087019 NM_001256398 c.G1373A p.G458E exonic . -

Hypomesus Transpacificus
Aquatic Toxicology 105 (2011) 369–377 Contents lists available at ScienceDirect Aquatic Toxicology jou rnal homepage: www.elsevier.com/locate/aquatox Sublethal responses to ammonia exposure in the endangered delta smelt; Hypomesus transpacificus (Fam. Osmeridae) ∗ 1 2 Richard E. Connon , Linda A. Deanovic, Erika B. Fritsch, Leandro S. D’Abronzo , Inge Werner Aquatic Toxicology Laboratory, Department of Anatomy, Physiology and Cell Biology, School of Veterinary Medicine, University of California, Davis, California 95616, United States a r t i c l e i n f o a b s t r a c t Article history: The delta smelt (Hypomesus transpacificus) is an endangered pelagic fish species endemic to the Received 9 May 2011 Sacramento-San Joaquin Estuary in Northern California, which acts as an indicator of ecosystem health Received in revised form 29 June 2011 in its habitat range. Interrogative tools are required to successfully monitor effects of contaminants upon Accepted 2 July 2011 the delta smelt, and to research potential causes of population decline in this species. We used microarray technology to investigate genome-wide effects in fish exposed to ammonia; one of multiple contami- Keywords: nants arising from wastewater treatment plants and agricultural runoff. A 4-day exposure of 57-day Hypomesus transpacificus + old juveniles resulted in a total ammonium (NH4 –N) median lethal concentration (LC50) of 13 mg/L, Delta smelt Microarray and a corresponding un-ionized ammonia (NH3) LC50 of 147 g/L. Using the previously designed delta + Biomarker smelt microarray we assessed altered gene transcription in juveniles exposed to 10 mg/L NH4 –N from Ammonia this 4-day exposure. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like,