A Grid Algorithm for High Throughput Fitting of Dose-Response Curve Data Yuhong Wang*, Ajit Jadhav, Noel Southal, Ruili Huang and Dac-Trung Nguyen
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Unsupervised Contour Representation and Estimation Using B-Splines and a Minimum Description Length Criterion Mário A
IEEE TRANSACTIONS ON IMAGE PROCESSING, VOL. 9, NO. 6, JUNE 2000 1075 Unsupervised Contour Representation and Estimation Using B-Splines and a Minimum Description Length Criterion Mário A. T. Figueiredo, Member, IEEE, José M. N. Leitão, Member, IEEE, and Anil K. Jain, Fellow, IEEE Abstract—This paper describes a new approach to adaptive having external/potential energy, which is a func- estimation of parametric deformable contours based on B-spline tion of certain features of the image The equilibrium (min- representations. The problem is formulated in a statistical imal total energy) configuration framework with the likelihood function being derived from a re- gion-based image model. The parameters of the image model, the (1) contour parameters, and the B-spline parameterization order (i.e., the number of control points) are all considered unknown. The is a compromise between smoothness (enforced by the elastic parameterization order is estimated via a minimum description nature of the model) and proximity to the desired image features length (MDL) type criterion. A deterministic iterative algorithm is (by action of the external potential). developed to implement the derived contour estimation criterion. Several drawbacks of conventional snakes, such as their “my- The result is an unsupervised parametric deformable contour: it adapts its degree of smoothness/complexity (number of control opia” (i.e., use of image data strictly along the boundary), have points) and it also estimates the observation (image) model stimulated a great amount of research; although most limitations parameters. The experiments reported in the paper, performed of the original formulation have been successfully addressed on synthetic and real (medical) images, confirm the adequacy and (see, e.g., [6], [9], [10], [34], [38], [43], [49], and [52]), non- good performance of the approach. -

Chapter 26: Mathcad-Data Analysis Functions
Lecture 3 MATHCAD-DATA ANALYSIS FUNCTIONS Objectives Graphs in MathCAD Built-in Functions for basic calculations: Square roots, Systems of linear equations Interpolation on data sets Linear regression Symbolic calculation Graphing with MathCAD Plotting vector against vector: The vectors must have equal number of elements. MathCAD plots values in its default units. To change units in the plot……? Divide your axis by the desired unit. Or remove the units from the defined vectors Use Graph Toolbox or [Shift-2] Or Insert/Graph from menu Graphing 1 20 2 28 time 3 min Temp 35 K time 4 42 Time min 5 49 40 40 Temp Temp 20 20 100 200 300 2 4 time Time Graphing with MathCAD 1 20 Plotting element by 2 28 element: define a time 3 min Temp 35 K 4 42 range variable 5 49 containing as many i 04 element as each of the vectors. 40 i:=0..4 Temp i 20 100 200 300 timei QuickPlots Use when you want to x 0 0.12 see what a function looks like 1 Create a x-y graph Enter the function on sin(x) 0 y-axis with parameter(s) 1 Enter the parameter on 0 2 4 6 x-axis x Graphing with MathCAD Plotting multiple curves:up to 16 curves in a single graph. Example: For 2 dependent variables (y) and 1 independent variable (x) Press shift2 (create a x-y plot) On the y axis enter the first y variable then press comma to enter the second y variable. On the x axis enter your x variable. -
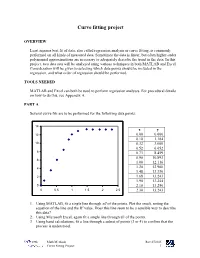
Curve Fitting Project
Curve fitting project OVERVIEW Least squares best fit of data, also called regression analysis or curve fitting, is commonly performed on all kinds of measured data. Sometimes the data is linear, but often higher-order polynomial approximations are necessary to adequately describe the trend in the data. In this project, two data sets will be analyzed using various techniques in both MATLAB and Excel. Consideration will be given to selecting which data points should be included in the regression, and what order of regression should be performed. TOOLS NEEDED MATLAB and Excel can both be used to perform regression analyses. For procedural details on how to do this, see Appendix A. PART A Several curve fits are to be performed for the following data points: 14 x y 12 0.00 0.000 0.10 1.184 10 0.32 3.600 0.52 6.052 8 0.73 8.459 0.90 10.893 6 1.00 12.116 4 1.20 12.900 1.48 13.330 2 1.68 13.243 1.90 13.244 0 2.10 13.250 0 0.5 1 1.5 2 2.5 2.30 13.243 1. Using MATLAB, fit a single line through all of the points. Plot the result, noting the equation of the line and the R2 value. Does this line seem to be a sensible way to describe this data? 2. Using Microsoft Excel, again fit a single line through all of the points. 3. Using hand calculations, fit a line through a subset of points (3 or 4) to confirm that the process is understood. -

Kwame Nkrumah University of Science and Technology, Kumasi
KWAME NKRUMAH UNIVERSITY OF SCIENCE AND TECHNOLOGY, KUMASI, GHANA Assessing the Social Impacts of Illegal Gold Mining Activities at Dunkwa-On-Offin by Judith Selassie Garr (B.A, Social Science) A Thesis submitted to the Department of Building Technology, College of Art and Built Environment in partial fulfilment of the requirement for a degree of MASTER OF SCIENCE NOVEMBER, 2018 DECLARATION I hereby declare that this work is the result of my own original research and this thesis has neither in whole nor in part been prescribed by another degree elsewhere. References to other people’s work have been duly cited. STUDENT: JUDITH S. GARR (PG1150417) Signature: ........................................................... Date: .................................................................. Certified by SUPERVISOR: PROF. EDWARD BADU Signature: ........................................................... Date: ................................................................... Certified by THE HEAD OF DEPARTMENT: PROF. B. K. BAIDEN Signature: ........................................................... Date: ................................................................... i ABSTRACT Mining activities are undertaken in many parts of the world where mineral deposits are found. In developing nations such as Ghana, the activity is done both legally and illegally, often with very little or no supervision, hence much damage is done to the water bodies where the activities are carried out. This study sought to assess the social impacts of illegal gold mining activities at Dunkwa-On-Offin, the capital town of Upper Denkyira East Municipality in the Central Region of Ghana. The main objectives of the research are to identify factors that trigger illegal mining; to identify social effects of illegal gold mining activities on inhabitants of Dunkwa-on-Offin; and to suggest effective ways in curbing illegal mining activities. Based on the approach to data collection, this study adopts both the quantitative and qualitative approach. -
Nonlinear Least-Squares Curve Fitting with Microsoft Excel Solver
Information • Textbooks • Media • Resources edited by Computer Bulletin Board Steven D. Gammon University of Idaho Moscow, ID 83844 Nonlinear Least-Squares Curve Fitting with Microsoft Excel Solver Daniel C. Harris Chemistry & Materials Branch, Research & Technology Division, Naval Air Warfare Center,China Lake, CA 93555 A powerful tool that is widely available in spreadsheets Unweighted Least Squares provides a simple means of fitting experimental data to non- linear functions. The procedure is so easy to use and its Experimental values of x and y from Figure 1 are listed mode of operation is so obvious that it is an excellent way in the first two columns of the spreadsheet in Figure 2. The for students to learn the underlying principle of least- vertical deviation of the ith point from the smooth curve is squares curve fitting. The purpose of this article is to intro- vertical deviation = yi (observed) – yi (calculated) (2) duce the method of Walsh and Diamond (1) to readers of = yi – (Axi + B/xi + C) this Journal, to extend their treatment to weighted least The least squares criterion is to find values of A, B, and squares, and to add a simple method for estimating uncer- C in eq 1 that minimize the sum of the squares of the verti- tainties in the least-square parameters. Other recipes for cal deviations of the points from the curve: curve fitting have been presented in numerous previous papers (2–16). n 2 Σ Consider the problem of fitting the experimental gas sum = yi ± Axi + B / xi + C (3) chromatography data (17) in Figure 1 with the van Deemter i =1 equation: where n is the total number of points (= 13 in Fig. -

(EU FP7 2013 - 612218/3D-NET) 3DNET Sops HOW-TO-DO PRACTICAL GUIDE
3D-NET (EU FP7 2013 - 612218/3D-NET) 3DNET SOPs HOW-TO-DO PRACTICAL GUIDE Curve fitting This is intended to be an aid-memoir for those involved in fitting biological data to dose response curves in why we do it and how to best do it. So why do we use non linear regression? Much of the data generated in Biology is sigmoidal dose response (E/[A]) curves, the aim is to generate estimates of upper and lower asymptotes, A50 and slope parameters and their associated confidence limits. How can we analyse E/[A] data? Why fit curves? What’s wrong with joining the dots? Fitting Uses all the data points to estimate each parameter Tell us the confidence that we can have in each parameter estimate Estimates a slope parameter Fitting curves to data is model fitting. The general equation for a sigmoidal dose-response curve is commonly referred to as the ‘Hill equation’ or the ‘four-parameter logistic equation”. (Top Bottom) Re sponse Bottom EC50hillslope 1 [Drug]hillslpoe The models most commonly used in Biology for fitting these data are ‘model 205 – Dose response one site -4 Parameter Logistic Model or Sigmoidal Dose-Response Model’ in XLfit (package used in ActivityBase or Excel) and ‘non-linear regression - sigmoidal variable slope’ in GraphPad Prism. What the computer does - Non-linear least squares Starts with an initial estimated value for each variable in the equation Generates the curve defined by these values Calculates the sum of squares Adjusts the variables to make the curve come closer to the data points (Marquardt method) Re-calculates the sum of squares Continues this iteration until there’s virtually no difference between successive fittings. -

Fitting Conics to Noisy Data Using Stochastic Linearization
Fitting Conics to Noisy Data Using Stochastic Linearization Marcus Baum and Uwe D. Hanebeck Abstract— Fitting conic sections, e.g., ellipses or circles, to LRF noisy data points is a fundamental sensor data processing problem, which frequently arises in robotics. In this paper, we introduce a new procedure for deriving a recursive Gaussian state estimator for fitting conics to data corrupted by additive Measurements Gaussian noise. For this purpose, the original exact implicit measurement equation is reformulated with the help of suitable Fig. 1: Laser rangefinder (LRF) scanning a circular object. approximations as an explicit measurement equation corrupted by multiplicative noise. Based on stochastic linearization, an efficient Gaussian state estimator is derived for the explicit measurement equation. The performance of the new approach distributions for the parameter vector of the conic based is evaluated by means of a typical ellipse fitting scenario. on Bayes’ rule. An explicit description of the parameter uncertainties with a probability distribution is in particular I. INTRODUCTION important for performing gating, i.e., dismissing unprobable In this work, the problem of fitting a conic section such measurements, and tracking multiple conics. Furthermore, as an ellipse or a circle to noisy data points is considered. the temporal evolution of the state can be captured with This fundamental problem arises in many applications related a stochastic system model, which allows to propagate the to robotics. A traditional application is computer vision, uncertainty about the state to the next time step. where ellipses and circles are fitted to features extracted A. Contributions from images [1]–[4] in order to detect, localize, and track objects. -

Small Molecule Screening Protocols
Small Molecule Screening Workshop July 29th, 2013 Sapporo, Japan Molecular Biophysics Group Structural Genomics Consortium University of Toronto On the cover: Cytosolic sulfotransferase family of proteins was screened against a library of 90 compounds by a thermodenaturation-based method (DSLS). Structures of cytosolic sulfotransferases are clustered according to their binding site structural similarity and surrounded by the molecules used in the binding experiments (Allali-Hassani et al., 2007, PLoS Biology. PMID: 17425406). Welcome to the workshop As part of the SGC’s ongoing program in structural genomics of human and malaria proteins we are also identifying small molecules that bind and stabilize our proteins, thereby promoting crystallization. We have implemented two screening platforms, based on either fluorimetry or static light scattering, to measure the increase in protein thermal stability upon binding of a ligand without the need to monitor enzyme activity (Vedadi et al (2006) PNAS. 103 (43):15835-40). Identification of small molecules that bind to proteins can provide valuable information on protein specificity, activity, as well as a practical means to facilitate 3D structure determination. This workshop aims to provide an opportunity for you to learn how to implement differential scanning fluorimetry (DSF) technique. We would like to take the opportunity to welcome you to our workshop. Masoud Vedadi Contact Information Masoud Vedadi, Ph.D. Principal Investigator, Molecular Biophysics Structural Genomics Consortium, Assistant Professor, Department of Pharmacology and Toxicology, University of Toronto, MaRS Centre, South Tower 101 College St.7 Floor,Room 714 Toronto, ON, M5G 1L7 Tel: 416 946 0897 E-mail: [email protected] www.sgc.utoronto.ca/biophysics Prepared by: Dr. -

Investigating Bias in the Application of Curve Fitting Programs To
Atmos. Meas. Tech., 8, 1469–1489, 2015 www.atmos-meas-tech.net/8/1469/2015/ doi:10.5194/amt-8-1469-2015 © Author(s) 2015. CC Attribution 3.0 License. Investigating bias in the application of curve fitting programs to atmospheric time series P. A. Pickers and A. C. Manning Centre for Ocean and Atmospheric Sciences, School of Environmental Sciences, University of East Anglia, Norwich, NR4 7TJ, UK Correspondence to: P. A. Pickers ([email protected]) Received: 31 May 2014 – Published in Atmos. Meas. Tech. Discuss.: 16 July 2014 Revised: 6 November 2014 – Accepted: 6 November 2014 – Published: 23 March 2015 Abstract. The decomposition of an atmospheric time series programs for certain types of data sets, and for certain types into its constituent parts is an essential tool for identifying of analyses and applications. In addition, we recommend that and isolating variations of interest from a data set, and is sensitivity tests are performed in any study using curve fitting widely used to obtain information about sources, sinks and programs, to ensure that results are not unduly influenced by trends in climatically important gases. Such procedures in- the input smoothing parameters chosen. volve fitting appropriate mathematical functions to the data. Our findings also have implications for previous studies However, it has been demonstrated that the application of that have relied on a single curve fitting program to inter- such curve fitting procedures can introduce bias, and thus in- pret atmospheric time series measurements. This is demon- fluence the scientific interpretation of the data sets. We inves- strated by using two other curve fitting programs to replicate tigate the potential for bias associated with the application of work in Piao et al. -
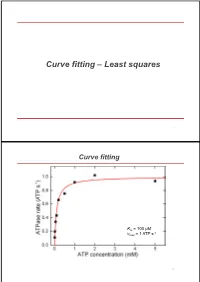
Curve Fitting – Least Squares
Curve fitting – Least squares 1 Curve fitting Km = 100 µM -1 vmax = 1 ATP s 2 Excurse error of multiple measurements Starting point: Measure N times the same parameter Obtained values are Gaussian distributed around mean with standard deviation σ What is the error of the mean of all measurements? Sum = x1 +x2 + ... + xN Variance of sum = N * σ2 (Central limit theorem) Standard deviation of sum N Mean = (x1 +x2 + ... + xN)/N N 1 Standard deviation of mean N (called standard error of mean) N N N 3 Excurse error propagation What is error for f(x,y,z,…) if we know errors of x,y,z,… (σx, σy, σz, …) for purely statistical errors? -> Individual variances add scaled by squared partial derivatives (if parameters are uncorrelated) Examples : Addition/substraction: squared errors add up squared relative Product: errors add up 4 Excurse error propagation squared relative Ratios: errors add up Powers: relative error times power Logarithms: error is relative error 5 Curve fitting – Least squares Starting point: - data set with N pairs of (xi,yi) - xi known exactly, -yi Gaussian distributed around true value with error σi - errors uncorrelated - function f(x) which shall describe the values y (y = f(x)) - f(x) depends on one or more parameters a [S] v vmax [S] KM 6 Curve fitting – Least squares 1 2 2 Probability to get y for given x [y i f (xi ;a)] / 2 i ] i i P(y i ;a) e 2 i [S] v vmax [S] KM 7 Curve fitting – Least squares N 1 2 2 Prob. -

Security Systems Services World Report
Security Systems Services World Report established in 1974, and a brand since 1981. www.datagroup.org Security Systems Services World Report Database Ref: 56162 This database is updated monthly. Security Systems Services World Report SECURITY SYSTEMS SERVICES WORLD REPORT The Security systems services Report has the following information. The base report has 59 chapters, plus the Excel spreadsheets & Access databases specified. This research provides World Data on Security systems services. The report is available in several Editions and Parts and the contents and cost of each part is shown below. The Client can choose the Edition required; and subsequently any Parts that are required from the After-Sales Service. Contents Description ....................................................................................................................................... 5 REPORT EDITIONS ........................................................................................................................... 6 World Report ....................................................................................................................................... 6 Regional Report ................................................................................................................................... 6 Country Report .................................................................................................................................... 6 Town & Country Report ...................................................................................................................... -

Characterization of Vixotrigine, a Broad-Spectrum Voltage-Gated Sodium Channel Blocker S
Supplemental material to this article can be found at: http://molpharm.aspetjournals.org/content/suppl/2021/04/02/99.1.49.DC1 1521-0111/99/1/49–59$35.00 https://doi.org/10.1124/molpharm.120.000079 MOLECULAR PHARMACOLOGY Mol Pharmacol 99:49–59, January 2021 Copyright ª 2020 by The Author(s) This is an open access article distributed under the CC BY-NC Attribution 4.0 International license. Characterization of Vixotrigine, a Broad-Spectrum Voltage-Gated Sodium Channel Blocker s Christopher A Hinckley, Yuri Kuryshev, Alissende Sers, Alexander Barre, Bruno Buisson, Himanshu Naik, and Mihaly Hajos1 Biogen, Cambridge, Massachusetts (C.A.H., H.N.); Charles River Laboratories Cleveland, Inc., Cleveland, Ohio (Y.K.); Neuroservice, Aix-en-Provence, France (A.S., A.B., B.B.); and Department of Comparative Medicine, Yale University, New Haven, Connecticut (M.H.) Received June 8, 2020; accepted October 15, 2020 Downloaded from ABSTRACT Voltage-gated sodium channels (Navs) are promising targets for profound state-dependent inhibition but a similar broad analgesic and antiepileptic therapies. Although specificity be- spectrum of action distinct from Nav1.7- and Nav1.8-specific tween Nav subtypes may be desirable to target specific neural blockers. We find that vixotrigine rapidly inhibits Navs and types, such as nociceptors in pain, many broadly acting Nav prolongs recovery from the fast-inactivated state. In native molpharm.aspetjournals.org inhibitors are clinically beneficial in neuropathic pain and rodent dorsal root ganglion sodium channels, we find that epilepsy. Here, we present the first systematic characterization vixotrigine shifts steady-state inactivation curves. Based on of vixotrigine, a Nav blocker.