2NCOMMS-18-06274-A Revise Supplementary Figure
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Genome-Scale Identification of Transcription Factors That Mediate An
ARTICLE DOI: 10.1038/s41467-018-04406-2 OPEN Genome-scale identification of transcription factors that mediate an inflammatory network during breast cellular transformation Zhe Ji 1,2,4, Lizhi He1, Asaf Rotem1,2,5, Andreas Janzer1,6, Christine S. Cheng2,7, Aviv Regev2,3 & Kevin Struhl 1 Transient activation of Src oncoprotein in non-transformed, breast epithelial cells can initiate an epigenetic switch to the stably transformed state via a positive feedback loop that involves 1234567890():,; the inflammatory transcription factors STAT3 and NF-κB. Here, we develop an experimental and computational pipeline that includes 1) a Bayesian network model (AccessTF) that accurately predicts protein-bound DNA sequence motifs based on chromatin accessibility, and 2) a scoring system (TFScore) that rank-orders transcription factors as candidates for being important for a biological process. Genetic experiments validate TFScore and suggest that more than 40 transcription factors contribute to the oncogenic state in this model. Interestingly, individual depletion of several of these factors results in similar transcriptional profiles, indicating that a complex and interconnected transcriptional network promotes a stable oncogenic state. The combined experimental and computational pipeline represents a general approach to comprehensively identify transcriptional regulators important for a biological process. 1 Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115, USA. 2 Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA. 3 Department of Biology, Howard Hughes Medical Institute and David H. Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA 20140, USA. 4Present address: Department of Pharmacology and Biomedical Engineering, Northwestern University, Evanston 60611 IL, USA. -

FOXK Transcription Factors Regulation and Critical Role in Cancer
Cancer Letters 458 (2019) 1–12 Contents lists available at ScienceDirect Cancer Letters journal homepage: www.elsevier.com/locate/canlet Mini-review FOXK transcription factors: Regulation and critical role in cancer T Ying Liua, Wei Dingb,HuGea,d, Murugavel Ponnusamya, Qiong Wangd, Xiaodan Haoa, Wei Wua, ∗∗ ∗ Yuan Zhanga, Wanpeng Yua, Xiang Aoa, , Jianxun Wanga,c, a Institute for Translational Medicine, College of Medicine, Qingdao University, Qingdao 266021, China b Department of Comprehensive Internal Medicine, Affiliated Hospital, Qingdao University, Qingdao 266003, China c School of Basic Medical Sciences, Qingdao University, Qingdao 266071, China d Molecular Informatics Department, Hengrui Pharmaceutical Co., Ltd., Shanghai 200245, China ARTICLE INFO ABSTRACT Keywords: Growing evidence suggests that alterations of gene expression including expression and activities of transcription FOXK1 factors are closely associated with carcinogenesis. Forkhead Box Class K (FOXK) proteins, FOXK1 and FOXK2, FOXK2 are a family of evolutionarily conserved transcriptional factors, which have recently been recognized as key ncRNAs transcriptional regulators involved in many types of cancer. Members of the FOXK family mediate a wide Biomarker spectrum of biological processes, including cell proliferation, differentiation, apoptosis, autophagy, cell cycle Therapeutic target progression, DNA damage and tumorigenesis. Therefore, the deregulation of FOXKs can affect the cell fate and they promote tumorigenesis as well as cancer progression. The mechanisms of FOXKs regulation including post- translational modifications (PTMs), microRNAs (miRNAs) and protein–protein interactions are well demon- strated. However, the detailed mechanisms of FOXKs activation and deregulation in cancer progression are still inconclusive. In this review, we summarize the regulatory mechanisms of FOXKs expression and activity, and their role in the development and progression of cancer. -

Using Liver Mirna Profiles to Predict Chemical Hepatocarcinogenesis”
“Using liver miRNA profiles to predict chemical hepatocarcinogenesis” By Costas Koufaris A thesis submitted to Imperial College London for the degree of Doctor of Philosophy Department of Surgery and Cancer Biomolecular Medicine Imperial College London April- 12011 - ABSTRACT Industrial, agricultural, and pharmaceutical requirements drive the development of a plethora of new chemical entities each year, many of which -for example drugs, pesticides, and food additives- have to be assessed for potential human health hazard. The current benchmark for risk assessement is the lifetime rodent bioassay which is expensive, time-consuming, laborious, requires the sacrifice of numerous animals, and is often irrelevant to humans. Hence alternative strategies to the rodent lifetime bioassay for prediction of chemical carcinogens are being pursued, especially for the liver which is an organ frequently affected by exogenous chemicals due to its detoxifying and metabolic roles. Numerous studies in recent years support the important role of microRNAs in cancer development, including hepatocellular carcinoma. The principal hypothesis of this project was that hepatic microRNA signatures can contribute to the earlier prediction of chemical hepatocarcinogens. Examination of livers from male Fischer rats treated with six chemical hepatocarcinogens, with diverse mode of actions for 90 days revealed that all the tested hepatocarcinogens affected the liver miRNAome from that early stage. Interestingly, a small set of microRNAs were identified whose expression -

FOXK2 Downregulation Suppresses EMT in Hepatocellular Carcinoma
Open Medicine 2020; 15: 702–708 Research Article Jian Kong#, Qingyun Zhang#, Xuefeng Liang, Wenbing Sun* FOXK2 downregulation suppresses EMT in hepatocellular carcinoma https://doi.org/10.1515/med-2020-0129 Keywords: hepatocellular carcinoma, FOXK2, epithe- received December 22, 2019; accepted March 31, 2020 lial–mesenchymal transition, Akt Abstract: Forkhead box K2 (FOXK2) was first identified as an NFAT-like interleukin-binding factor. FOXK2 has been reported to act as either oncogene or tumor suppressor. However, functional and regulating mechan- 1 Introduction isms of FOXK2 in epithelial–mesenchymal transition ( ) (EMT) in hepatocellular carcinoma (HCC) remain un- Hepatocellular carcinoma HCC is listed as the sixth clear. An FOXK2-specific siRNA was employed to most common neoplasm and the third leading cause of decrease the endogenous expression of FOXK2. MTT cancer death, which has been recognized as a major [ ] assay, colony formation and transwell assay were used cause of death among cirrhotic patients 1 . Surgical to evaluate proliferation, migration and invasion of resection, liver transplantation and radiofrequency [ ] Hep3B and HCCLM3 cells, respectively. The protein ablation are considered potentially curative for HCC 2 . expression associated with EMT and Akt signaling However, most HCC patients are diagnosed at advanced pathways was evaluated using western blot. FOXK2 stages when surgical treatments are unsuitable. Only downregulation could inhibit cell proliferation and sorafenib and regorafenib are proven to prolong survival [ – ] colony formation and suppress migration and invasion in HCC 3 8 . Therefore, development of new targeted in Hep3B and HCCLM3 cells. The expression of E- therapy for HCC treatment is urgent. ( ) cadherin was significantly upregulated, and the expres- The structure of Forkhead box FOX proteins contains fi sion of snail and p-Akt was significantly downregulated conserved DNA binding domain, which is de ned as the - - in siFOXK2-transfected cells compared with control cells. -

Hepatic Posttranscriptional Network Comprised of CCR4–NOT Deadenylase and FGF21 Maintains Systemic Metabolic Homeostasis
Hepatic posttranscriptional network comprised of CCR4–NOT deadenylase and FGF21 maintains systemic metabolic homeostasis Masahiro Moritaa,b,c,1,2, Nadeem Siddiquid,e,1, Sakie Katsumuraa,b, Christopher Rouyad,e, Ola Larssonf, Takeshi Nagashimag, Bahareh Hekmatnejadh,i, Akinori Takahashij, Hiroshi Kiyonarik, Mengwei Zanga,b, René St-Arnaudh,i, Yuichi Oikel, Vincent Giguèred,e,m, Ivan Topisirovicd,m,n, Mariko Okada-Hatakeyamag,o, Tadashi Yamamotoj,2, and Nahum Sonenbergd,e,2 aDepartment of Molecular Medicine, University of Texas Health Science Center at San Antonio, San Antonio, TX 78229; bBarshop Institute for Longevity and Aging Studies, University of Texas Health Science Center at San Antonio, San Antonio, TX 78229; cInstitute of Resource Development and Analysis, Kumamoto University, 860-0811 Kumamoto, Japan; dDepartment of Biochemistry, McGill University, Montreal, QC H3A 1A3, Canada; eGoodman Cancer Research Centre, McGill University, Montreal, QC H3A 1A3, Canada; fDepartment of Oncology-Pathology, Scilifelab, Karolinska Institutet, SE-171 76 Stockholm, Sweden; gLaboratory for Integrated Cellular Systems, RIKEN Center for Integrative Medical Sciences, Yokohama, 230-0045 Kanagawa, Japan; hResearch Centre, Shriners Hospital for Children–Canada, Montreal, QC H4A 0A9, Canada; iDepartment of Human Genetics, McGill University, Montreal, QC H3A 2T5, Canada; jCell Signal Unit, Okinawa Institute of Science and Technology Graduate University, Onna-son, 904-0495 Okinawa, Japan; kLaboratories for Animal Resource Development and Genetic Engineering, RIKEN Center for Biosystems Dynamics Research, Kobe, 650-0047 Hyogo, Japan; lDepartment of Molecular Genetics, Graduate School of Medical Sciences, Kumamoto University, 860-8556 Kumamoto, Japan; mGerald Bronfman Department of Oncology, McGill University, Montreal, QC H2W 1S6, Canada; nLady Davis Institute for Medical Research, Sir Mortimer B. -

Content Based Search in Gene Expression Databases and a Meta-Analysis of Host Responses to Infection
Content Based Search in Gene Expression Databases and a Meta-analysis of Host Responses to Infection A Thesis Submitted to the Faculty of Drexel University by Francis X. Bell in partial fulfillment of the requirements for the degree of Doctor of Philosophy November 2015 c Copyright 2015 Francis X. Bell. All Rights Reserved. ii Acknowledgments I would like to acknowledge and thank my advisor, Dr. Ahmet Sacan. Without his advice, support, and patience I would not have been able to accomplish all that I have. I would also like to thank my committee members and the Biomed Faculty that have guided me. I would like to give a special thanks for the members of the bioinformatics lab, in particular the members of the Sacan lab: Rehman Qureshi, Daisy Heng Yang, April Chunyu Zhao, and Yiqian Zhou. Thank you for creating a pleasant and friendly environment in the lab. I give the members of my family my sincerest gratitude for all that they have done for me. I cannot begin to repay my parents for their sacrifices. I am eternally grateful for everything they have done. The support of my sisters and their encouragement gave me the strength to persevere to the end. iii Table of Contents LIST OF TABLES.......................................................................... vii LIST OF FIGURES ........................................................................ xiv ABSTRACT ................................................................................ xvii 1. A BRIEF INTRODUCTION TO GENE EXPRESSION............................. 1 1.1 Central Dogma of Molecular Biology........................................... 1 1.1.1 Basic Transfers .......................................................... 1 1.1.2 Uncommon Transfers ................................................... 3 1.2 Gene Expression ................................................................. 4 1.2.1 Estimating Gene Expression ............................................ 4 1.2.2 DNA Microarrays ...................................................... -

Unique Epigenetic Programming Distinguishes Regenerative Spermatogonial Stem Cells in the Developing Mouse Testis Affiliations A
bioRxiv preprint doi: https://doi.org/10.1101/674457; this version posted June 17, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Unique Epigenetic Programming Distinguishes Regenerative Spermatogonial Stem Cells in the Developing Mouse Testis Authors: Keren Cheng1, I-Chung Chen1, Benjamin J. Hale2, Brian P. Hermann1, Christopher B. Geyer2, Jon M. Oatley3, and John R. McCarrey1,* Affiliations 1Department of Biology, University of Texas at San Antonio, San Antonio, TX 2 Department of Anatomy and Cell Biology at the Brody School of Medicine and East Carolina Diabetes and Obesity Institute, East Carolina University, Greenville, NC 3Center for Reproductive Biology, School of Molecular Biosciences, College of Veterinary Medicine, Washington State University, Pullman, WA *Correspondence: [email protected] Abstract Spermatogonial stem cells (SSCs) both self-renew and give rise to progenitor spermatogonia that enter steady-state spermatogenesis in the mammalian testis. However, questions remain regarding the extent to which SSCs and progenitors represent stably distinct spermatogonial subtypes. Here we provide the first multiparametric integrative analysis of mammalian germ cell epigenomes comparable to that done by the ENCODE Project for >100 somatic cell types. Differentially expressed genes distinguishing SSCs and progenitors showed distinct histone bioRxiv preprint doi: https://doi.org/10.1101/674457; this version posted June 17, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. modification patterns as well as differences in distal intergenic low-methylated regions. -

Discerning the Role of Foxa1 in Mammary Gland
DISCERNING THE ROLE OF FOXA1 IN MAMMARY GLAND DEVELOPMENT AND BREAST CANCER by GINA MARIE BERNARDO Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy Dissertation Adviser: Dr. Ruth A. Keri Department of Pharmacology CASE WESTERN RESERVE UNIVERSITY January, 2012 CASE WESTERN RESERVE UNIVERSITY SCHOOL OF GRADUATE STUDIES We hereby approve the thesis/dissertation of Gina M. Bernardo ______________________________________________________ Ph.D. candidate for the ________________________________degree *. Monica Montano, Ph.D. (signed)_______________________________________________ (chair of the committee) Richard Hanson, Ph.D. ________________________________________________ Mark Jackson, Ph.D. ________________________________________________ Noa Noy, Ph.D. ________________________________________________ Ruth Keri, Ph.D. ________________________________________________ ________________________________________________ July 29, 2011 (date) _______________________ *We also certify that written approval has been obtained for any proprietary material contained therein. DEDICATION To my parents, I will forever be indebted. iii TABLE OF CONTENTS Signature Page ii Dedication iii Table of Contents iv List of Tables vii List of Figures ix Acknowledgements xi List of Abbreviations xiii Abstract 1 Chapter 1 Introduction 3 1.1 The FOXA family of transcription factors 3 1.2 The nuclear receptor superfamily 6 1.2.1 The androgen receptor 1.2.2 The estrogen receptor 1.3 FOXA1 in development 13 1.3.1 Pancreas and Kidney -

PDF-Document
Supplementary Material Investigating the role of microRNA and Transcription Factor co-regulatory networks in Multiple Sclerosis pathogenesis Nicoletta Nuzziello1, Laura Vilardo2, Paride Pelucchi2, Arianna Consiglio1, Sabino Liuni1, Maria Trojano3 and Maria Liguori1* 1National Research Council, Institute of Biomedical Technologies, Bari Unit, Bari, Italy 2National Research Council, Institute of Biomedical Technologies, Segrate Unit, Milan, Italy 3Department of Basic Sciences, Neurosciences and Sense Organs, University of Bari, Bari, Italy Supplementary Figure S1 Frequencies of GO terms and canonical pathways. (a) Histogram illustrates the GO terms associated to assembled sub-networks. (b) Histogram illustrates the canonical pathways associated to assembled sub-network. a b Legends for Supplementary Tables Supplementary Table S1 List of feedback (FBL) and feed-forward (FFL) loops in miRNA-TF co-regulatory network. Supplementary Table S2 List of significantly (adj p-value < 0.05) GO-term involved in MS. The first column (from the left) listed the GO-term (biological processes) involved in MS. For each functional class, the main attributes (gene count, p-value, adjusted p-value of the enriched terms for multiple testing using the Benjamini correction) have been detailed. In the last column (on the right), we summarized the target genes involved in each enriched GO-term. Supplementary Table S3 List of significantly (adj p-value < 0.05) enriched pathway involved in MS. The first column (from the left) listed the enriched pathway involved in MS. For each pathway, the main attributes (gene count, p-value, adjusted p-value of the enriched terms for multiple testing using the Benjamini correction) have been detailed. In the last column (on the right), we summarized the target genes involved in each enriched pathway. -

Foxk1 and Foxk2 in Insulin Regulation of Cellular and Mitochondrial Metabolism
ARTICLE https://doi.org/10.1038/s41467-019-09418-0 OPEN FoxK1 and FoxK2 in insulin regulation of cellular and mitochondrial metabolism Masaji Sakaguchi1,2,3, Weikang Cai1,2, Chih-Hao Wang1,2, Carly T. Cederquist1,2, Marcos Damasio1,2, Erica P. Homan1,2, Thiago Batista1,2, Alfred K. Ramirez1,2, Manoj K. Gupta1,2, Martin Steger4, Nicolai J. Wewer Albrechtsen4,5,6, Shailendra Kumar Singh7, Eiichi Araki3, Matthias Mann 4, Sven Enerbäck 8 & C. Ronald Kahn1,2 1234567890():,; A major target of insulin signaling is the FoxO family of Forkhead transcription factors, which translocate from the nucleus to the cytoplasm following insulin-stimulated phosphorylation. Here we show that the Forkhead transcription factors FoxK1 and FoxK2 are also downstream targets of insulin action, but that following insulin stimulation, they translocate from the cytoplasm to nucleus, reciprocal to the translocation of FoxO1. FoxK1/FoxK2 translocation to the nucleus is dependent on the Akt-mTOR pathway, while its localization to the cytoplasm in the basal state is dependent on GSK3. Knockdown of FoxK1 and FoxK2 in liver cells results in upregulation of genes related to apoptosis and down-regulation of genes involved in cell cycle and lipid metabolism. This is associated with decreased cell proliferation and altered mito- chondrial fatty acid metabolism. Thus, FoxK1/K2 are reciprocally regulated to FoxO1 following insulin stimulation and play a critical role in the control of apoptosis, metabolism and mitochondrial function. 1 Sections of Integrative Physiology and Metabolism and Islet Cell Biology and Regenerative Medicine, Joslin Diabetes Center, Boston, MA 02215, USA. 2 Department of Medicine, Harvard Medical School, Boston, MA 02215, USA. -
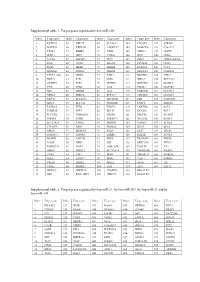
Target Genes Regulated by Hsa-Mir-21, by Hsa-Mir-203, by Hsa-Mir-21 and by Hsa-Mir-143
Supplemental table 1: Target genes regulated by hsa-miR-205 Index Target gene Index Target gene Index Target gene Index Target gene Index Target gene 1 KCTD20 35 UBE2Z 69 SLC38A1 103 LPCAT1 137 STK38L 2 MAPK14 36 YWHAH 70 ANGPTL7 104 MARCKS 138 C1orf123 3 TXNL1 37 RBBP4 71 CTGF 105 MED13 139 GUCD1 4 SPDL1 38 LRP1 72 CYR61 106 IPO7 140 CDK6 5 TCF20 39 IMPAD1 73 TP73 107 PHC2 141 CDKN2AIPNL 6 RAN 40 GNAS 74 EGLN2 108 PICALM 142 CLIP1 7 RGS6 41 MED1 75 ERBB2 109 PLAGL2 143 CUL5 8 HOXA11 42 INPPL1 76 PRRG4 110 NDUFA4 144 C6orf201 9 PAPPA-AS1 43 DDX5 77 F2RL2 111 NDUFB2 145 VTI1A 10 PRR15 44 E2F1 78 GOT1 112 NIPA2 146 SLC5A12 11 ACTRT3 45 E2F5 79 NUFIP2 113 NOTCH2 147 MAML2 12 YES1 46 ZEB2 80 IL24 114 PANK1 148 MAP3K9 13 SRC 47 ERBB3 81 IL32 115 PARD6B 149 NUDT21 14 NPRL3 48 PRKCE 82 RNF217 116 TMEM66 150 DNAJA1 15 NFAT5 49 SLC41A1 83 ZNF585B 117 EZR 151 CCDC108 16 XPOT 50 SLC7A2 84 SIGMAR1 118 ENPP4 152 SHISA6 17 KCTD16 51 ZEB1 85 VEGFA 119 LRRTM4 153 ACP1 18 TMSB4X 52 PHF8 86 BCL9L 120 KCNJ10 154 BCL2 19 PLCXD2 53 TMEM201 87 CREB1 121 PHLPP2 155 NCAPG 20 TNFSF8 54 PTPRJ 88 SERINC3 122 YEATS2 156 KLHL5 21 SLC25A25 55 ETNK1 89 HMGB3 123 VAMP1 157 ACSL4 22 C11orf74 56 XPR1 90 SRD5A1 124 RTN3 158 BCL6 23 GM2A 57 MRPL44 91 PTEN 125 RFX7 159 ITGA5 24 SMNDC1 58 TM9SF2 92 ESRRG 126 RAP2B 160 ACSL1 25 BAMBI 59 PAIP2B 93 PRLR 127 TRAF3IP1 161 EID2B 26 LCOR 60 NEK9 94 ICK 128 SERTAD2 162 TEX35 27 TMEM239 61 NOX5 95 LOH12CR1 129 TOLLIP 163 YY1 28 AMOT 62 DMXL2 96 SLC39A14 130 TMEM55B 164 SMAD1 29 CDK1 63 ETF1 97 BDP1 131 TMEM123 165 SMAD4 30 SQLE 64