Isolation and Characterization of Bliai, an Isoschizomer of Clai from Bacillus Licheniformis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Restriction Endonucleases
Molecular Biology Problem Solver: A Laboratory Guide. Edited by Alan S. Gerstein Copyright © 2001 by Wiley-Liss, Inc. ISBNs: 0-471-37972-7 (Paper); 0-471-22390-5 (Electronic) 9 Restriction Endonucleases Derek Robinson, Paul R. Walsh, and Joseph A. Bonventre Background Information . 226 Which Restriction Enzymes Are Commercially Available? . 226 Why Are Some Enzymes More Expensive Than Others? . 227 What Can You Do to Reduce the Cost of Working with Restriction Enzymes? . 228 If You Could Select among Several Restriction Enzymes for Your Application, What Criteria Should You Consider to Make the Most Appropriate Choice? . 229 What Are the General Properties of Restriction Endonucleases? . 232 What Insight Is Provided by a Restriction Enzyme’s Quality Control Data? . 233 How Stable Are Restriction Enzymes? . 236 How Stable Are Diluted Restriction Enzymes? . 236 Simple Digests . 236 How Should You Set up a Simple Restriction Digest? . 236 Is It Wise to Modify the Suggested Reaction Conditions? . 237 Complex Restriction Digestions . 239 How Can a Substrate Affect the Restriction Digest? . 239 Should You Alter the Reaction Volume and DNA Concentration? . 241 Double Digests: Simultaneous or Sequential? . 242 225 Genomic Digests . 244 When Preparing Genomic DNA for Southern Blotting, How Can You Determine If Complete Digestion Has Been Obtained? . 244 What Are Your Options If You Must Create Additional Rare or Unique Restriction Sites? . 247 Troubleshooting . 255 What Can Cause a Simple Restriction Digest to Fail? . 255 The Volume of Enzyme in the Vial Appears Very Low. Did Leakage Occur during Shipment? . 259 The Enzyme Shipment Sat on the Shipping Dock for Two Days. -

Bpuami: a Novel Saci Neoschizomer from Bacillus Pumilus Discovered in an Isolate from Amazon Basin, Recognizing 5'-Gag↓Ctc-3'
Brazilian Journal of Microbiology (2006) 37:96-100 ISSN 1517-8382 BPUAMI: A NOVEL SACI NEOSCHIZOMER FROM BACILLUS PUMILUS DISCOVERED IN AN ISOLATE FROM AMAZON BASIN, RECOGNIZING 5'-GAG↓CTC-3' Jocelei M. Chies1,2,*; Ana C. de O. Dias1; Hélio M. M. Maia3; Spartaco Astolfi-Filho2,3 1Centro de Biotecnologia, Universidade Federal do Rio Grande do Sul, RS, Brasil; 2Curso de Pós-Graduação em Biologia Molecular, Universidade de Brasília, Brasília, DF, Brasil; 3Centro de Apoio Multidisciplinar, Universidade Federal do Amazonas, Manaus, AM, Brasil Submitted: June 27, 2005; Returned to authors for corrections: November 16, 2005; Approved: January 12, 2006 ABSTRACT A strain of Bacillus pumilus was isolated and identified from water samples collected from a small affluent of the Amazon River. Type II restriction endonuclease activity was detected in these bacteria. The enzyme was purified and the molecular weight of the native protein estimated by gel filtration and SDS-PAGE. The optimum pH, temperature and salt requirements were determined. Quality control assays showed the complete absence of “nonspecific nucleases.” Restriction cleavage analysis and DNA sequencing of restriction fragments allowed the unequivocal demonstration of 5´GAG↓CTC3´ as the recognition sequence. This enzyme was named BpuAmI and is apparently a neoschizomer of the prototype restriction endonuclease SacI. This is the first report of an isoschizomer and/or neoschizomer of the prototype SacI identified in the genus Bacillus. Key words: type II restriction endonuclease, BpuAmI, SacI, neoschizomers, Bacillus pumilus INTRODUCTION thousands of taxonomically diverse bacteria for enzymes with new characteristics. However, to our knowledge, there has Restriction endonucleases are enzymes which recognize not been any report to date of an isoschizomer and/or short DNA sequences and cleave DNA in both strands. -

1. Restriction Endonucleases (® Fermentas 2006)
We guarantee that these products are Introduction ..........................................................................................................2 free of contaminating activities. Restriction Endonucleases Guide Our stringent quality control with the most FastDigest™ Restriction Endonucleases .........................................................2 Fermentas Restriction Endonucleases............................................................3 advanced tests guarantees you pure 1 Alphabetic List of Commercially Available Restriction Endonucleases.........6 products for your experiments. ISO9001 Recognition Specifi cities...............................................................................16 and ISO14001 is your assurance of con- Commercial Restriction Enzymes Sorted by the Type of DNA Ends Generated sistency and lot-to-lot reproducibility. Enzymes Generating 5’-protruding Ends.......................................................18 Enzymes Generating 3’-protruding Ends.......................................................18 ™ PureExtreme Quality will provide the Enzymes Generating Blunt Ends...................................................................19 performance you need for your most Enzymes Cleaving DNA on the Both Sides of Their Recognition Sequences.....19 demanding experiments. PureExtreme™ Quality PureExtreme™ Quality Guarantee..................................................................20 Activity Assay.................................................................................................20 -

Restriction Endonucleases
Restriction Endonucleases TECHNICAL GUIDE UPDATE 2017/18 be INSPIRED drive DISCOVERY stay GENUINE RESTRICTION ENZYMES FROM NEB Cut Smarter with Restriction Enzymes from NEB® Looking to bring CONVENIENCE to your workflow? Simplify Reaction Setup and Double Activity of DNA Modifying Enzymes in CutSmart Buffer: Digestion with CutSmart® Buffer Clone Smarter! Activity Enzyme Required Supplements Over 210 restriction enzymes are 100% active in a single buffer, in CutSmart Phosphatases: CutSmart Buffer, making it significantly easier to set up your Alkaline Phosphatase (CIP) + + + double digest reactions. Since CutSmart Buffer includes BSA, there Antarctic Phosphatase + + + Requires Zn2+ Quick CIP + + + are fewer tubes and pipetting steps to worry about. Additionally, Shrimp Alkaline Phosphatase (rSAP) + + + many DNA modifying enzymes are 100% active in CutSmart Ligases: T4 DNA Ligase + + + Requires ATP Buffer, eliminating the need for subsequent purification. E. coli DNA Ligase + + + Requires NAD T3 DNA Ligase + + + Requires ATP + PEG For more information, visit www.NEBCutSmart.com T7 DNA Ligase + + + Requires ATP + PEG Polymerases: T4 DNA Polymerase + + + DNA Polymerase I, Large (Klenow) Frag. + + + DNA Polymerase I + + + DNA Polymerase Klenow Exo– + + + Bst DNA Polymerase + + + ™ phi29 DNA Polymerase + + + Speed up Digestions with Time-Saver T7 DNA Polymerase (unmodified) + + + Qualified Restriction Enzymes Transferases/Kinases: T4 Polynucleotide Kinase + + + Requires ATP + DTT T4 PNK (3´ phosphatase minus) + + + Requires ATP + DTT > 190 of our restriction enzymes are able to digest DNA in CpG Methyltransferase (M. SssI) + + + 5–15 minutes, and can safely be used overnight with no loss of GpC Methyltransferase (M. CviPI) + Requires DTT T4 Phage β-glucosyltransferase + + + sample. For added convenience and flexibility, most of these are Nucleases, other: supplied with CutSmart Buffer. -
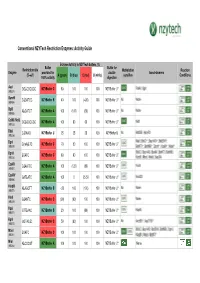
Conventional Nzytech Restriction Enzymes: Activity Guide
Conventional NZYTech Restriction Enzymes: Activity Guide Enzyme Activity in NZYTech buffers (%) Buffer Buffer for Restriction site Methylation Reaction Enzyme provided for double Isoschizomers (5’ →→→3’) A (green) B (blue) C (red) U (white) sensitive Conditions 100% activity digestion AscI GG ↓↓↓CGCGCC NZYBuffer C 80 100 100 100 NZYBuffer U * PalAI, SgsI (MB231) BamHI G↓↓↓GATCC NZYBuffer B 40 100 (<20) 100 NZYBuffer U * No None (MB064) BglII A↓↓↓GATCT NZYBuffer A 100 (100) (60) 100 NZYBuffer U * No None (MB065) CciNI (NotI) GC ↓↓↓GGCCGC NZYBuffer A 100 80 60 100 NZYBuffer U * NotI (MB153) DdeI C↓↓↓TNAG NZYBuffer U 25 25 25 100 NZYBuffer U No BstDEI, HpyF3I (MB236) MalI, BfuCI ♦♦♦, Bsp143I ♦♦♦, BstENII ♦♦♦, DpnI G(mA)↓↓↓TC NZYBuffer C 70 50 100 100 NZYBuffer U * (MB078) BstKTI ♦♦♦, BstMBI ♦♦♦, DpnII ♦♦♦, Kzo9I ♦♦♦, NdeII ♦♦♦ BfuCI, Bsp143I, BssMI, BstKTI, BstMBI, DpnII ↓↓↓GATC NZYBuffer C 80 80 100 100 NZYBuffer U * (MB233) Kzo9I, MboI, NdeII, Sau3AI EcoRI G↓↓↓AATTC NZYBuffer A 100 (120) (80) 100 NZYBuffer U * FunII (MB067) EcoRV GAT ↓↓↓ATC NZYBuffer A 100 0 25-50 100 NZYBuffer U * Eco32I (MB068) HindIII A↓↓↓AGCTT NZYBuffer B <20 100 (100) 100 NZYBuffer U * No None (MB070) HinfI G↓↓↓ANTC NZYBuffer C (90) (90) 100 100 NZYBuffer U * None (MB239) HpaI GTT ↓↓↓AAC NZYBuffer B 20 100 (80) 100 NZYBuffer U * KspAI (MB071) KpnI GGTAC ↓↓↓C NZYBuffer C 50 (80) 100 100 NZYBuffer U * No Acc65I ♦♦♦, Asp718I ♦♦♦ (MB072) BfuCI, Bsp143I, BstENII, BstMBI, DpnII, MboI ↓↓↓GATC NZYBuffer C 100 100 100 100 NZYBuffer U * (MB241) Kzo9II, NdeII, BstKTI ♦♦♦ MluI -

Biology of DNA Restriction THOMAS A
MICROBIOLOGICAL REVIEWS, June 1993, p. 434-450 Vol. 57, No. 2 0146-0749/93/020434-17$02.00/0 Copyright © 1993, American Society for Microbiology Biology of DNA Restriction THOMAS A. BICKLE'* AND DETLEV H. KRUGER2 Department ofMicrobiology, Biozentrum, Basel University, Klingelbergstrasse 70, CH-4056 Basel, Switzerland, 1 and Institute of Virology, Charite School ofMedicine, Humboldt University, D-0-1040 Berlin, Gernany2 INTRODUCTION ........................................................................ 434 TYPE I R-M SYSTEMS ........................................................................ 434 Type I Systems Form Families of Related Enzymes ...................................................................435 Structure of hsd Genes ........................................................................ 435 Evolution of DNA Sequence Recognition by Recombination between hsdS Genes .........................***436 Mutations Affecting Modification Activity........................................................................ 437 TYPE II R-M SYSTEMS........................................................................ 437 Evolutionary Aspects ........................................................................ 437 Control of Expression of Type II RM Genes .....................................................438 Cytosine Can Be Methylated on Either C-5 Or NA: Consequences for Mutagenesis...............438 Type II Restriction Endonucleases That Require Two Recognition Sites for Cleavage.439 What Is the Function of Type IIS Enzymes.440 -

Curriculum Vitae SIR RICHARD JOHN ROBERTS ADDRESS PERSONAL
Curriculum Vitae SIR RICHARD JOHN ROBERTS ADDRESS New England Biolabs 240 County Road, Ipswich, MA 02138 USA Email: [email protected] Telephone: (978) 380-7405 / Fax: (978) 380-7406 PERSONAL Born on September 6, 1943, Derby, England EDUCATION 1962-1965 University of Sheffield, Sheffield, England B.Sc. in Chemistry 1966-1968 University of Sheffield, Sheffield, England Ph.D. in Organic Chemistry POSITIONS 2005- Chief Scientific Officer, New England Biolabs 1992-2005 Research Director, New England Biolabs 1986-92 Assistant Director for Research, Cold Spring Harbor Laboratory 1972-86 Senior Staff Investigator, Cold Spring Harbor Laboratory 1971-1972 Research Associate in Biochemistry, Harvard University 1969-1970 Research Fellow, Harvard University OUTSIDE ACTIVITIES 1974-1992 Consultant and Chairman of Scientific Advisory Board New England Biolabs 1977-1985 Scientific Advisory Board, Genex Corp. 1977-1987 Editorial Board: Nucleic Acids Research 1979-1984 Editorial Board: Journal of Biological Chemistry 1982-1989 Member: National Advisory Committee of GENBANK 1984-1986 Member: National Advisory Committee of BIONET 1985-1988 Panel member: NIH Study Section in Biochemistry. 1985-2002 Editorial Board: Bioinformatics (formerly CABIOS) 1987-1990 Chairman: National Advisory Committee of BIONET 1987-2009 Senior Executive Editor: Nucleic Acids Research 1990-1992 Panel member: NCI Cancer Centers Support Grant Review Committee 1993-1995 Panel member: NLM Study Section/Comp. Biol. 1994-2000 Scientific Advisory Board, Molecular Tool 1994- Patron of the Oxford International Biomedical Center 1996-1998 Visiting Professor, University of Bath, UK. 1996-2000 Chairman, NCI Board of Scientific Counselors 1996-1999 Scientific Advisory Board, Oxford Molecular Group 1997-2001 Editorial Board: Current Opinion Chem. Biol. -

Restriction Enzymes Are Molecular Scissors
Molecular Scissors Restriction enzymes are molecular scissors DR. ABUL HASAN SARDAR Assistant Professor and Head Department of Microbiology Sarsuna College RESTRICTION ENZYMES • A restriction enzyme (or restriction endonuclease) is an enzyme that cuts double- stranded or single stranded DNA at specific recognition nucleotide sequences known as restriction sites. Property of restriction enzymes • They that link adjacent nucleotides in DNA molecules. HOW RESTRICTION ENZYMES WORKS? • Restriction enzymes recognize a specific sequence of nucleotides, and produce a double-stranded cut in the DNA, these cuts are of two types: • BLUNT ENDS. • STICKY ENDS. Blunt end Sticky end BLUNT ENDS • These blunt ended fragments can be joined to any other DNA fragment with blunt ends. • Enzymes useful for certain types of DNA cloning experiments “STICKY ENDS” ARE USEFUL DNA fragments with complimentary sticky ends can be combined to create new molecules which allows the creation and manipulation of DNA sequences from different sources. • While recognition sequences vary widely , with lengths between 4 and 8 nucleotides, many of them are palindromic. PALINDROMES IN DNA SEQUENCES Genetic palindromes are similar to verbal palindromes. A palindromic sequence in DNA is one in which the 5’ to 3’ base pair sequence is identical on both strands (the 5’ and 3’ ends refers to the chemical structure of the DNA). PALINDROME SEQUENCES • The mirror like palindrome in which the same forward and backwards are on a single strand of DNA strand, as in GTAATG • The Inverted repeat palindromes is also a sequence that reads the same forward and backwards, but the forward and backward sequences are found in complementary DNA strands (GTATAC being complementary to CATATG) • Inverted repeat palindromes are more common and have greater biological importance than mirror- like palindromes. -

Anza Restriction Enzyme Cloning System Complete, One-Buffer System— for Beautifully Simple Cloning Cloning Has Never Been Simpler
Anza Restriction Enzyme Cloning System Complete, one-buffer system— for beautifully simple cloning Cloning has never been simpler Finally, forget the frustrations of finding compatible buffers and sorting through protocols for your restriction enzyme digests—start getting reliable results for your downstream experiments. The Invitrogen™ Anza™ Restriction Enzyme Cloning System is a complete system, comprised of: DNA modifying enzymes 128 restriction enzymes + 5 DNA modifying enzymes Anza T4 DNA Ligase Master Mix Anza Thermosensitive All Anza™ restriction enzymes work together cohesively and are fully functional Alkaline Phosphatase with the single Anza™ buffer. Anza T4 Polynucleotide Kinase The system offers: Anza DNA Blunt End Kit • One buffer for all restriction enzymes Anza DNA End Repair Kit • One digestion protocol for all DNA types • Complete digestion in 15 minutes • Overnight digestion without star activity Convenient buffer formats All Anza restriction enzymes come with an Anza 10X Buffer and an Anza 10X Red Buffer to give you the flexibility you require. The red buffer includes a density reagent containing red and yellow tracking dyes that migrate with 800 bp DNA fragments and faster than the 10 bp DNA fragments, respectively, in a 1% agarose gel. This eliminates tedious dye addition steps prior to gel loading and is compatible with downstream applications.* * For applications that require analysis by fluorescence excitation, Anza 10X Buffer is recommended, as the Anza 10X Red Buffer may interfere with some fluorescence measurements. 2 A one-buffer system Eliminate the frustration of trying to find compatible buffers for all your enzymes. All Anza restriction enzymes allow for complete digestion using a single Anza buffer. -

2019-20 NEB Catalog Technical Reference
Reference Appendix Technical Support – Featured Tools & for scientists, by scientists Resources As a partner to the scientific community, New England Biolabs is committed to Optimizing Restriction Enzyme Reactions providing top quality tools and scientific expertise. This philosophy still stands, 290 and has led to long-standing relationships with many of our fellow scientists. Performance Chart for NEB's commitment to scientists is the same regardless of whether or not they 293 Restriction Enzymes purchase product from NEB: their ongoing research is supported by our catalog, Troubleshooting Guide website and technical staff. 349 for Cloning NEB's technical support model is unique as it utilizes most of the scientists at NEB. Several of our product lines have designated technical support scientists 334 Methylation Sensitivity assigned to servicing customers in those application areas. Any questions regarding a product could be dealt with by one of the technical support Guidelines for scientists, the product manager who manufactures it, the product development 337 PCR Optimization scientist who optimizes it, or a researcher who uses the product in their daily Cleavage Close to the research. As such, customers are supported by scientists and often experts in 343 End of DNA Fragments the product or its application. To access technical support: 288 Online Interactive Tools Call 1-800-632-7799 (Monday – Friday: 9:00 am - 6:00 pm EST) Submit an online form at www.neb.com/techsupport View NEB TV Episode #22 Email [email protected] to learn more about our Technical Support program. International customers can contact a local NEB subsidiary or distributor. For more information see inside back cover. -

Review Type II Restriction Endonucleases
CMLS, Cell. Mol. Life Sci. 62 (2005) 685–707 1420-682X/05/060685-23 DOI 10.1007/s00018-004-4513-1 CMLS Cellular and Molecular Life Sciences © Birkhäuser Verlag, Basel, 2005 Review Type II restriction endonucleases: structure and mechanism A. Pingouda,*, M. Fuxreiterb, V.Pingouda and W.Wendea a Institut für Biochemie, Justus-Liebig-Universität, Heinrich-Buff-Ring 58, 35392 Giessen (Germany), Fax: +49 641 9935409, e-mail: [email protected] b Institute of Enzymology, Biological Research Centre, Hungarian Academy of Sciences, Karolina ut 29, 1113 Budapest (Hungary) Received 15 November 2004; accepted 9 December 2004 Abstract. Type II restriction endonucleases are compo- few restriction endonucleases discovered thus far do not nents of restriction modification systems that protect belong to the PD…D/ExK family of enzymes, but rather bacteria and archaea against invading foreign DNA. Most have active sites typical of other endonuclease families. are homodimeric or tetrameric enzymes that cleave DNA The present review deals with new developments in the at defined sites of 4–8 bp in length and require Mg2+ ions field of Type II restriction endonucleases. One of the for catalysis. They differ in the details of the recognition more interesting aspects is the increasing awareness of process and the mode of cleavage, indicators that these the diversity of Type II restriction enzymes. Nevertheless, enzymes are more diverse than originally thought. Still, structural studies summarized herein deal with the more most of them have a similar structural core and seem to common subtypes. A major emphasis of this review will share a common mechanism of DNA cleavage, suggest- be on target site location and the mechanism of catalysis, ing that they evolved from a common ancestor. -

Gel Electrophoresis Applications Gel Electrophoresis Applications
Restriction Enzymes (also known as restriction endonucleases) What are restriction enzymes? • An enzyme that cuts DNA at specific nucleotide sequences known as restriction sites • Naturally found in bacteria and have evolved to provide a defense mechanism against invading viruses • In bacteria, restriction enzymes selectively cut up foreign DNA in a process called restriction Restriction Enzymes in Bacteria Restriction Enzymes • Cut both strands of the DNA double helix • Over 3000 enzymes have been identified • More than 600 available commercially • Routinely used for DNA modification and manipulation in laboratories (“molecular scissors”) Restriction Sites • Also known as recognition sites • Generally genetic palindromic sequences • A palindromic sequence in DNA one in which the 5’ to 3’ base pair sequence is identical on both strands • Usually a 4 or 6 base pair sequence Restriction Sites • The enzymes scan DNA sequences, find a very specific set of nucleotides and make a cut Hae III • HaeIII is a restriction enzyme that searches the DNA molecule until it finds this sequence of four nitrogen bases - GGCC 5’ TGACGGGTTCGAGGCCAG 3’ 3’ ACTGCCCAAGGTCCGGTC 5’ Hae III • Once the recognition site was found HaeIII will go to work cutting (cleaving) the DNA Blunt Ends versus Sticky Ends • Hae III produces “blunt ends” when cleaving DNA • Other enzymes produce “sticky ends” blunt end sticky end Restriction Enzyme Names • Named after the type of bacteria in which the enzyme is found and the order in which the restriction enzyme was identified and isolated EcoRI for example R strain of E.coli bacteria I as it is was the first E. coli restriction enzyme to be discovered.