(Syrmaticus Reevesii) Lysozyme
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hybridization & Zoogeographic Patterns in Pheasants
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Paul Johnsgard Collection Papers in the Biological Sciences 1983 Hybridization & Zoogeographic Patterns in Pheasants Paul A. Johnsgard University of Nebraska-Lincoln, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/johnsgard Part of the Ornithology Commons Johnsgard, Paul A., "Hybridization & Zoogeographic Patterns in Pheasants" (1983). Paul Johnsgard Collection. 17. https://digitalcommons.unl.edu/johnsgard/17 This Article is brought to you for free and open access by the Papers in the Biological Sciences at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Paul Johnsgard Collection by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. HYBRIDIZATION & ZOOGEOGRAPHIC PATTERNS IN PHEASANTS PAUL A. JOHNSGARD The purpose of this paper is to infonn members of the W.P.A. of an unusual scientific use of the extent and significance of hybridization among pheasants (tribe Phasianini in the proposed classification of Johnsgard~ 1973). This has occasionally occurred naturally, as for example between such locally sympatric species pairs as the kalij (Lophura leucol11elana) and the silver pheasant (L. nycthelnera), but usually occurs "'accidentally" in captive birds, especially in the absence of conspecific mates. Rarely has it been specifically planned for scientific purposes, such as for obtaining genetic, morphological, or biochemical information on hybrid haemoglobins (Brush. 1967), trans ferins (Crozier, 1967), or immunoelectrophoretic comparisons of blood sera (Sato, Ishi and HiraI, 1967). The literature has been summarized by Gray (1958), Delacour (1977), and Rutgers and Norris (1970). Some of these alleged hybrids, especially those not involving other Galliformes, were inadequately doculnented, and in a few cases such as a supposed hybrid between domestic fowl (Gallus gal/us) and the lyrebird (Menura novaehollandiae) can be discounted. -

Copper Pheasant
Bird Research News Vol.3 No.11 2006.11.10. Last Revised:2013.11.28. Copper Pheasant Yamadori (Jpn) Syrmaticus soemmerringii Morphology and classification bly smaller white part in the boundary zone between the two sub- species ranges (Kawaji 2004). Confusion between the two subspe- cies poses a problem to game hunters and government officials Classification: Galliformes Phasianidae concerned every year because ijimae has been removed from game Wing length: ♂ 205-230mm ♀ 192-219mm bird list since 1979 due to the population decline. Since subspecies Tail length: ♂ 415-952mm ♀ 164-205mm classification could become a major issue in conservation and Culmen length: ♂ 23-31mm ♀ 22-26mm social scenes in the future, genetic analyses are currently underway Tarsus length: ♂ 57-69mm ♀ 53-60mm (Sakanashi et al. 2006). Weight: ♂ 943-1348g ♀ 745-1000g Habitat: All measurement values are of subspecies Syrmaticus soemmerringii scintillans after Kiyosu (1978). Copper Pheasants are a forest dweller, generally preferring a broad -leaved forest. But they also occur in conifer plantations, such as Appearance: Japanese cedar and cypress. They occasionally bathe in the sun Plumage colors of Copper Pheasants and dust in grasslands of a cutover. vary between subspecies. In general, however, male is reddish brown on the Life history upper and underparts with black, chest- nut, yellow and white lateral bars in the 123456789101112 long tail (Photo 1). In the subspecies of breeding season non-breeding season the southern Japan (S.s. soemmerringii and ijimae), the plumage is generally Breeding system: darker with dark red brown and black Copper Pheasants are assumed to be polygamous like Common horizontal bars in the tail. -

CHAPTER 5 PRODUCTS of ANIMAL ORIGIN, NOT ELSEWHERE SPECIFIED OR INCLUDED I 5-1 Notes: 1
)&f1y3X CHAPTER 5 PRODUCTS OF ANIMAL ORIGIN, NOT ELSEWHERE SPECIFIED OR INCLUDED I 5-1 Notes: 1. This chapter does not cover: (a) Edible products (other than guts, bladders and stomachs of animals, whole and pieces thereof, and animal blood, liquid or dried); (b) Hides or skins (including furskins) other than goods of heading 0505 and parings and simlar waste of raw hides or skins of heading 0511 (chapter 41 or 43); (c) Animal textile materials, other than horsehair and horsehair waste (section XI); or (d) Prepared knots or tufts for broom or brush making (heading 9603). 2. For the purposes of heading 0501, the sorting of hair by length (provided the root ends and tip ends, respectively, are not arranged together) shall be deemed not to constitute working. 3. Throughout the tariff schedule, elephant, walrus, narwhal and wild boar tusks, rhinoceros horns and the teeth of all animals are regarded as "ivory." 4. Throughout the tariff schedule, the expression "horsehair" means hair of the manes or tails of equine or bovine animals. Additional U.S. Note 1. (a) Except as provided in paragraphs (b) and (c) of this note, the importation of the feathers or skin of any bird is hereby prohibited. Such prohibition shall apply to the feathers or skin of any bird: (i) Whether raw or processed; (ii) Whether the whole plumage or skin or any part of either; (iii) Whether or not attached to a whole bird or any part thereof; and (iv) Whether or not forming part of another article. (b) Paragraph (a) shall not apply: (i) In respect of any of the following birds (other than any such bird which, whether or not raised in captivity, is a wild bird): chickens (including hens and roosters), turkeys, guineas, geese, ducks, pigeons, ostriches, rheas, English ring-necked pheasants and pea fowl; (ii) To any importation for scientific or educational purposes; (iii) To the importation of fully manufactured artificial flies used for fishing; (iv) To the importation of birds which are classifiable under subheading 9804.00.55; and (v) To the importation of live birds. -

Phylogenetic Relationships of the Phasianidae Reveals Possible Non-Pheasant Taxa
Journal of Heredity 2003:94(6):472–489 Ó 2003 The American Genetic Association DOI: 10.1093/jhered/esg092 Phylogenetic Relationships of the Phasianidae Reveals Possible Non-Pheasant Taxa K. L. BUSH AND C. STROBECK From the Department of Biological Sciences, University of Alberta–Edmonton, Edmonton, Alberta T6G 2E9, Canada. Address correspondence to Krista Bush at the address above, or e-mail: [email protected]. Abstract The phylogenetic relationships of 21 pheasant and 6 non-pheasant species were determined using nucleotide sequences from the mitochondrial cytochrome b gene. Maximum parsimony and maximum likelihood analysis were used to try to resolve the phylogenetic relationships within Phasianidae. Both the degree of resolution and strength of support are improved over previous studies due to the testing of a number of species from multiple pheasant genera, but several major ambiguities persist. Polyplectron bicalcaratum (grey peacock pheasant) is shown not to be a pheasant. Alternatively, it appears ancestral to either the partridges or peafowl. Pucrasia macrolopha macrolopha (koklass) and Gallus gallus (red jungle fowl) both emerge as non-pheasant genera. Monophyly of the pheasant group is challenged if Pucrasia macrolopha macrolopha and Gallus gallus are considered to be pheasants. The placement of Catreus wallichii (cheer) within the pheasants also remains undetermined, as does the cause for the great sequence divergence in Chrysolophus pictus obscurus (black-throated golden). These results suggest that alterations in taxonomic -
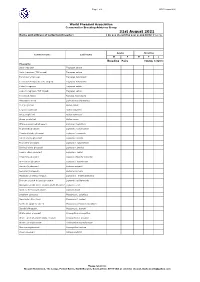
31St August 2021 Name and Address of Collection/Breeder: Do You Closed Ring Your Young Birds? Yes / No
Page 1 of 3 WPA Census 2021 World Pheasant Association Conservation Breeding Advisory Group 31st August 2021 Name and address of collection/breeder: Do you closed ring your young birds? Yes / No Adults Juveniles Common name Latin name M F M F ? Breeding Pairs YOUNG 12 MTH+ Pheasants Satyr tragopan Tragopan satyra Satyr tragopan (TRS ringed) Tragopan satyra Temminck's tragopan Tragopan temminckii Temminck's tragopan (TRT ringed) Tragopan temminckii Cabot's tragopan Tragopan caboti Cabot's tragopan (TRT ringed) Tragopan caboti Koklass pheasant Pucrasia macrolopha Himalayan monal Lophophorus impeyanus Red junglefowl Gallus gallus Ceylon junglefowl Gallus lafayettei Grey junglefowl Gallus sonneratii Green junglefowl Gallus varius White-crested kalij pheasant Lophura l. hamiltoni Nepal Kalij pheasant Lophura l. leucomelana Crawfurd's kalij pheasant Lophura l. crawfurdi Lineated kalij pheasant Lophura l. lineata True silver pheasant Lophura n. nycthemera Berlioz’s silver pheasant Lophura n. berliozi Lewis’s silver pheasant Lophura n. lewisi Edwards's pheasant Lophura edwardsi edwardsi Vietnamese pheasant Lophura e. hatinhensis Swinhoe's pheasant Lophura swinhoii Salvadori's pheasant Lophura inornata Malaysian crestless fireback Lophura e. erythrophthalma Bornean crested fireback pheasant Lophura i. ignita/nobilis Malaysian crestless fireback/Vieillot's Pheasant Lophura i. rufa Siamese fireback pheasant Lophura diardi Southern Cavcasus Phasianus C. colchicus Manchurian Ring Neck Phasianus C. pallasi Northern Japanese Green Phasianus versicolor -

Recognizing Decorative Body Feathers of Pheasants and Related Birds
Citation: Trail, P.W. 2006. Recognizing Decorative Body Feathers of Pheasants and Related Birds. Identification Guides for Wildlife Law Enforcement No. 9. USFWS, National Fish and Wildlife Forensics Laboratory, Ashland, OR. Recognizing Decorative Body Feathers of Pheasants and Related Birds Identification Guides for Wildlife Law Enforcement No. 9 Pepper W. Trail U.S. Fish and Wildlife Service National Fish and Wildlife Forensics Laboratory 1490 East Main Street Ashland, OR 97520 [email protected] July 2006 The colorful body feathers of chickens, pheasants, guineafowl, and related birds are often used to decorate earrings, hair ties, dreamcatchers, and other small crafted items. None of these species are native to North America, and none are listed under the Endangered Species Act or CITES. Familiarity with these feathers will allow Special Agents and Wildlife Inspectors to recognize the legal use of decorative feathers and to avoid unnecessary seizures. The birds frequently used for such decorative items include the following: • Chicken (Gallus gallus) • Indian Peafowl (Pavo cristatus) • Ring-necked Pheasant (Phasianus colchicus) • Golden Pheasant (Chrysolophus pictus) • Lady Amherst Pheasant (Chrysolophus amherstiae) • Reeve’s Pheasant (Syrmaticus reevesii) • Silver Pheasant (Lophura nycthemera) • Helmeted Guineafowl (Numida meleagris) • Vulturine Guineafowl (Acryllium vulturinum) In the following pages, an assortment of body feathers from each of these species is illustrated. Many male pheasants exhibit a variety of different colors and patterns on their body feathers, and an attempt has been made to illustrate the range of types for each species. In addition, the many domestic breeds of chickens vary greatly in plumage, and so chicken feathers are very diverse in appearance, as shown. -

New Sites for Mrs Hume's Pheasant Syrmaticus Humiae in North-East
Forktail 21 (2005) SHORT NOTES 183 New sites for Mrs Hume’s Pheasant Syrmaticus humiae in north-east India based on hunters’ specimens and local reports ANWARUDDIN CHOUDHURY Mrs Hume’s Pheasant Syrmaticus humiae is a poorly DISTRIBUTION known globally threatened (Vulnerable) species (BirdLife International 2004). It is thinly distributed in I observed the species at three previously known sites the hill tracts of north-eastern India, north and west (Shiroi, Murlen and Phawngpui), recorded 20 new Myanmar, south-west China and north Thailand (Ali sites based on live captured birds or preserved speci- and Ripley 1987, Fuller and Garson 2000, Han Lian- mens in villages, and identified an additional 24 new xian 1997).Very little current information on its status sites where villagers reported the species (Appendix, and distribution is available (Fuller and Garson 2000, Fig. 1). In Nagaland, BirdLife International (2001) BirdLife International 2001). Recent fieldwork in north-east India prior to this study had resulted in few records (Katju 1996, Kaul et al. 1996, Choudhury 1997, 1998, 2000, 2001, Robson 1999). No previous survey had specifically targeted the species. I carried out surveys in Nagaland, Manipur and Mizoram during 1996–2004 to assess the current distribution and status of the species, mainly from hunters’ specimens and local reports. METHODS I carried out surveys in: Nagaland in June 1996, January, February, April and October 2001, February 2002 and February 2004; Manipur in January 1996, January 2001, October 2001 and February 2002; and Mizoram in April 2000 and February 2001. These states are almost entirely mountainous. The climate is monsoonal with hot wet summers and cool dry winters (although winter rains are also not uncommon); annual rainfall is 1,000–6,000 mm. -

A Multigene Phylogeny of Galliformes Supports a Single Origin of Erectile Ability in Non-Feathered Facial Traits
J. Avian Biol. 39: 438Á445, 2008 doi: 10.1111/j.2008.0908-8857.04270.x # 2008 The Authors. J. Compilation # 2008 J. Avian Biol. Received 14 May 2007, accepted 5 November 2007 A multigene phylogeny of Galliformes supports a single origin of erectile ability in non-feathered facial traits Rebecca T. Kimball and Edward L. Braun R. T. Kimball (correspondence) and E. L. Braun, Dept. of Zoology, Univ. of Florida, P.O. Box 118525, Gainesville, FL 32611 USA. E-mail: [email protected] Many species in the avian order Galliformes have bare (or ‘‘fleshy’’) regions on their head, ranging from simple featherless regions to specialized structures such as combs or wattles. Sexual selection for these traits has been demonstrated in several species within the largest galliform family, the Phasianidae, though it has also been suggested that such traits are important in heat loss. These fleshy traits exhibit substantial variation in shape, color, location and use in displays, raising the question of whether these traits are homologous. To examine the evolution of fleshy traits, we estimated the phylogeny of galliforms using sequences from four nuclear loci and two mitochondrial regions. The resulting phylogeny suggests multiple gains and/or losses of fleshy traits. However, it also indicated that the ability to erect rapidly the fleshy traits is restricted to a single, well-supported lineage that includes species such as the wild turkey Meleagris gallopavo and ring-necked pheasant Phasianus colchicus. The most parsimonious interpretation of this result is a single evolution of the physiological mechanisms that underlie trait erection despite the variation in color, location, and structure of fleshy traits that suggest other aspects of the traits may not be homologous. -

Dramatic Decline of the Vulnerable Reeves's Pheasant Syrmaticus Reevesii, Endemic to Central China
Dramatic decline of the Vulnerable Reeves’s pheasant Syrmaticus reevesii, endemic to central China C HUNFA Z HOU,JILIANG X U and Z HENGWANG Z HANG Abstract The current status and distribution of the regions (Zheng & Wang, 1998; Collar et al., 2001). Reeves’s Vulnerable Reeves’s pheasant Syrmaticus reevesii, endemic pheasant is sensitive to expansion of development and there to central China, is poorly known. To obtain updated is no space for the species to shift its range in central China, information on its status we selected 89 candidate sites in six especially with the high degree of habitat fragmentation in provinces and one municipality in central China and this area (Zheng & Wang, 1998). Previous surveys (Xu et al., conducted interviews and field surveys from April 2011 to 1996; Ma et al., 2009) have indicated declines in the April 2012. Interviews demonstrated the pheasant has distribution and population density of Reeves’s pheasant disappeared from 46% of the surveyed sites. Our results but these surveys did not cover the entire range of the also revealed a population decline at 46 sites, including species or lacked accurate data. protected areas, although population densities in protected In the study reported here we conducted a large-scale areas were higher than those in non-protected areas. Eighty- and intensive survey of Reeves’s pheasant from April 2011 three, 26 and 20% of the surveyed sites had evidence of to April 2012. We aimed to: (1) ascertain the current poaching, habitat loss and use of poison, respectively, which distribution and population status of Reeves’s pheasant in were the three major threats to this species. -

Gallus Gallus Spadiceus) in Oil Palm Plantation Habitat, Malaysia
Pakistan J. Zool., vol. 43(5), pp. 833-840, 2011. Variation in Home Range Size Exhibited by Red Junglefowl (Gallus gallus spadiceus) in Oil Palm Plantation Habitat, Malaysia Muhammad Irshad Arshad* 1 and Mohamed Zakaria2 1College of Agriculture, Dera Ghazi Khan. 2Faculty of Forestry, University Putra Malaysia, 43400 UPM Serdang, Selangor, Malaysia Abstract.- A Radio telemetry study on Red Junglefowl (Gallus gallus spadiceus) was conducted in oil palm (Elaeis guineensis) plantation at Sungai Sedu Estate, Selangor, Malaysia from October 1996 to July 1997. The main objective of the study was to examine the ranging behaviour of the species. Four Red Junglefowls (3 males and 1 female) were caught using decoy and leg trap method. They were then equipped with single stage 16 g transmitters and were radio-tracked using Mariner 57 receiver. The radiolocation was taken every 30 minutes by triangulation. The results show that the daily and monthly home range size of male was greater than that of a female. Similarly the home range size of a male without a female was greater than with a female. Environmental factors such as temperature, relative humidity, sunshine duration and cloud cover have no effect on the size of home range. The movement (distance travelled) contributes 49.1% of the variability on home range size. The total daily movement of male was greater than that of a female. The Red Junglefowl travelled more in the morning than in the afternoon and evening. In general, the size of home range varies according to several factors such as when the male is establishing and defending its territory. -

2Xotlc ~Heagahtg 3
2xotlc ~heagahtg 3. Adequate nutritious diet. Most of the species listed as endan eaptiVe/PltVu1.7ement iV1d ~'to?a7t1tion gered or threatened are receivin.g attention in captivity and success IS by Mickey Dllson, Glendale, AZ being attained with many of these. (Aug/Sept 1979 ) ;11iLket{ OliSOl1 has dOl1e mO'le There are 16 genera comprising 48 Edwards' Pheasant The Edwards' Pheasant Lophura nJo'lk 011 pheasMts Md othe'l tjAlLi recognized species of pheasants. In some species there are numerous sub edwardsi is a small and beautiful bird 11aceous 6i'lds thM almost t:lJ1lfol1e species or races bringing the number first discovered in 1895 with a small else i11 Ame'l£CM atJ£cultu'le. d-le of forms to about 150. distribution in central Vietnam near Hue (Way). It was first imported in has alJValfs 6eel1 a J1JiLLiJ1'j tea.che'l, All of the pheasants, including peafowl and junglefowl, are found in 1925 to France by Dr. Jean Delacour, Sha'li115 his k110J1Jl£d5e a60ut these Asia except the Congo Peacock of when he brought some 15 individuals 6i'lds and he J1Jas a pwnee'l in 'lep'lO Africa. to his collection. It was successfully propagated and distributed to zoos ~nd duci115 mAAlf o~ the mO'le di~~£cuLt During the past 50 years radical changes have taken place in much of aviculturists in Europe and Amenca. species. Asia which have severely decimated But recently it has become increasingly wild populations of many species of difficult to breed and abnormalities in he pheasants are among pheasants. -

Pheasants of Mizoram (India): Present Status of Diversity and Distribution
www.sciencevision.org Sci Vis 11 (4), 218-223 October-December, 2011 Original Research ISSN (print) 0975-6175 ISSN (online) 2229-6026 Pheasants of Mizoram (India): Present status of diversity and distribution H. Lalthanzara, Vanramliana and Lalramliana Department of Zoology, Pachhunga University College, Aizawl 796001, India Received 23 December 2011 | Accepted 29 December2011 ABSTRACT Preliminary survey on the diversity and distribution of pheasants (Phasianidae: Galliformes) was conducted for 20 months (May 2010 - December 2011) in the state of Mizoram, northeast India. Field survey at important protected areas and collection of secondary information’s indicated that six species of pheasants are present in Mizoram (i.e. 11.8% of the world pheasant species). They are Green Peafowl (Pavo muticus Linnaeus, Mizo - Ârawn), Hume’s Pheasant (Syrmaticus humiae Hume, Mizo - Vavu), Blyth’s Tragopan (Tragopan blythii Jerdon, Mizo - Vangâ), Red Junglefowl (Gallus gallus Linnaeus, Mizo - Ramâr), Kalij Pheasant (Lophura leucomelananos Latham, Mizo - Vahrit) and Grey Peacock Pheasant (Polyplectron bicalcaratum Linnaeus, Mizo - Varihaw). There is only one endangered species i.e. P. muticus, while the vulnerable species T. blythii is recorded at two protected areas. S. humiae is a near threatened species; population of this species is thinly dis- tributed in eastern side of Mizoram along/near the Myanmar border. The three lower risk categories of pheasants (G. gallus, L. leucomelanos and P. bicalcaratum) are resident species, found in most parts of the state. L. leucomelanos is the most common species, found in all 11 protected areas fol- lowed by the G. gallus and P. bicalcaratum, both of them are found in 9 protected areas.