2019 RISE Symposium Abstract Book
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A COMPARATIVE ANALYSIS of SONG and RESPONSES to SONG PLAYBACK University of Colorado at Denver
FINAL REPORT A COMPARATIVE ANALYSIS OF SONG AND RESPONSES TO SONG PLAYBACK IN THE AVIAN GENUS PIPILO Peter S. Kaplan Department of Psychology University of Colorado at Denver Abstract An experiment was undertaken to characterize the responses of Green-tailed Towhees (Pipilo chlorura) and Rufous-sided Towhees (P. erythophthalmus) to each others' songs and to the songs of five other towhee species, plus one hybrid form. A total of 12 Green-Tailed Towhees and 10 Rufous-sided Towhees from Boulder and Gilpin counties were studied at three field sites: the Doudy Draw Trail, the National Center for Atmospheric Research (NCAR), and on private land at the mouth of Coal Creek Canyon. In May, each bird was mist-netted and banded to facilitate individual identification. During the subsequent playback phase in June and July, each individual received one three-part playback trial on each of 7 consecutive or near-consecutive days. A 9-min playback trial consisted of a 3-min "pre-play" period, during which the bird was observed in the absence of song playback, a 3-min "play" period, in which tape recorded song was played to the subject from a central point in his territory, and a 3-min "post-play" period when the bird • was again observed in the absence of song playback. Order of presentation of song exemplars from different towhee species were randomized across birds. The main dependent measure was the change in the number of songs produced by the subject bird during song playback, relative to the pre-play period. Results showed that Green-tailed Towhees responded by significantly increasing their rate of singing, but only in response to Green-tailed Towhee songs. -

21 Sep 2018 Lists of Victims and Hosts of the Parasitic
version: 21 Sep 2018 Lists of victims and hosts of the parasitic cowbirds (Molothrus). Peter E. Lowther, Field Museum Brood parasitism is an awkward term to describe an interaction between two species in which, as in predator-prey relationships, one species gains at the expense of the other. Brood parasites "prey" upon parental care. Victimized species usually have reduced breeding success, partly because of the additional cost of caring for alien eggs and young, and partly because of the behavior of brood parasites (both adults and young) which may directly and adversely affect the survival of the victim's own eggs or young. About 1% of all bird species, among 7 families, are brood parasites. The 5 species of brood parasitic “cowbirds” are currently all treated as members of the genus Molothrus. Host selection is an active process. Not all species co-occurring with brood parasites are equally likely to be selected nor are they of equal quality as hosts. Rather, to varying degrees, brood parasites are specialized for certain categories of hosts. Brood parasites may rely on a single host species to rear their young or may distribute their eggs among many species, seemingly without regard to any characteristics of potential hosts. Lists of species are not the best means to describe interactions between a brood parasitic species and its hosts. Such lists do not necessarily reflect the taxonomy used by the brood parasites themselves nor do they accurately reflect the complex interactions within bird communities (see Ortega 1998: 183-184). Host lists do, however, offer some insight into the process of host selection and do emphasize the wide variety of features than can impact on host selection. -

Life History Account for California Towhee
California Wildlife Habitat Relationships System California Department of Fish and Wildlife California Interagency Wildlife Task Group CALIFORNIA TOWHEE Melozone crissalis Family: EMBERIZIDAE Order: PASSERIFORMES Class: AVES B484 Written by: D. Dobkin, S. Granholm Reviewed by: L. Mewaldt Edited by: R. Duke Updated by: CWHR Program Staff, November 2014 DISTRIBUTION, ABUNDANCE, AND SEASONALITY The former brown towhee recently has been split into the California towhee and the canyon towhee, M. fusca (American Ornithologists' Union 1989). The California towhee is a common, characteristic resident of foothills and lowlands in most of cismontane California. Frequents open chaparral and coastal scrub, as well as brush-land patches in open riparian, hardwood hardwood-conifer, cropland, and urban habitats. Commonly uses edges of dense chaparral and brushy edges of densely wooded habitats. Also occurs in lowest montane habitats of similar structure in southern California, and locally in Siskiyou and western Modoc cos. Local on coastal slope north of southern Humboldt Co., and apparently absent from western San Joaquin Valley (Grinnell and Miller 1944, McCaskie et al. 1979, Garrett and Dunn 1981). The Inyo California towhee, M. c. eremophilus, occurs only in the Argus Mountains of southwestern Inyo Co. SPECIFlC HABITAT REQUIREMENTS Feeding: Feeds on seeds, insects, and some fruits. Gleans and scratches in litter, picks seeds and fruits from plants, and rarely flycatches (Davis 1957). Prefers to forage on open ground adjacent to brushy cover. Insects are important in breeding season, often constituting a third of the diet (Martin et al. 1961). Cover: Shrubs in broken chaparral, margins of dense chaparral, willow thickets, and brushy understory of open wooded habitats provide cover. -

Voice in Communication and Relationships Among Brown Towhees
THE CONDOR VOLUME 66 SEPTEMBER-OCTOBER, 1964 NUMBER 5 VOICE IN COMMUNICATION AND RELATIONSHIPS AMONG BROWN TOWHEES By JOET.MARSHALL,JR. This paper seeks to answer two questions: (1) What is the function of each song and call in brown towhees; that is, what information does a bird communicate to its fellows vocally, o’r how does it regulate their behavior by its voice? (2) What evi- dence does voice offer for understanding relationship by descent within the closely-knit group of brown towhee species? For the first, I would extend the analysis of Quain- tance (1938,194l) to all members of the group. As to the second question, an ingenious evolutionary reconstruction, based on museum and habitat studies, has been developed by Davis (1951). Do vocal attributes agree with his scheme? The three speciesof brown towhees, genus Pipdo, are the same size and general color and are more similar to each other than any one of them is to other ground-inhabiting finches in the same genus and in the genus Melozorte. Indeed, so close is their relation- ship that the same calls can easily be discerned in each species; although differing in timbre, similarity in form and usage proclaims them to be homologous. The Abert Towhee (Pipdo abed) occupies dense riparian woodland and mesquite thickets of the Colorado River and Gila River drainages, mostly in Arizona. The Brown Towhee proper (Pip20 fuscus) lives in brushy margins of openings in the southwestern United States and Mexico. The White-throated Towhee (Pipilo aZbicoZZis)inhabits brushy slopes, often with tree yuccas, in Puebla and Oaxaca, MCxico. -
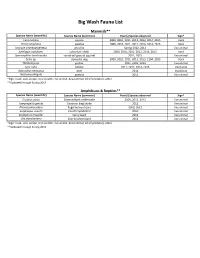
Flora and Fauna List.Xlsx
Big Wash Fauna List Mammals** Species Name (scientific) Species Name (common) Year(s) Species observed Sign* Canis latrans coyote 2009, 2010, 2011, 2012, 2013, 2014, 2015 track Pecari angulatus javalina 2009, 2010, 2011, 2012, 2013, 2014, 2015 track Urocyon cinereoargenteus gray fox Spring 2012, 2013 live animal Sylvilagus audubonii cottontail rabbit 2009, 2010, 2011, 2012, 2013, 2015 track Spermophilus tereticaudus round‐tail ground squirrel 2011, 2015 live animal Canis sp. domestic dog 2009, 2010, 2011, 2012, 2013, 2104, 2015 track Thomomys sp. gopher 2011, 2102, 2013 live animal Lynx rufus bobcat 2011, 2012, 2014, 2015 track/scat Odocoileus hemionus deer 2011 track/scat Neotoma albigula packrat 2011 live animal *Sign: track, scat, scrape, tree scratch, live animal, dead animal, kill (if predator), other **Updated through Spring 2015 Amphibians & Reptiles** Species Name (scientific) Species Name (common) Year(s) Species observed Sign* Crotalus atrox Diamondback rattlesnake 2009, 2012, 2013 live animal Lampropeltis getula Common king Snake 2012 live animal Phrynosoma solare Regal horned lizard 2009, 2012 live animal Scaphiopus couchii Couch's Spadefoot 2010 live animal Sceloporus magister Spiny lizard 2013 live animal Uta stansburiana Side‐bloched lizard 2013 live animal *Sign: track, scat, scrape, tree scratch, live animal, dead animal, kill (if predator), other **Updated through Spring 2015 Birds*** Species Name (scientific) Species Name (common) Year(s) Species observed Sign* Melozone aberti Abert's Towhee 2009, 2010, 2011, 2012, -

Behavioral, Morphological, and Ecological Trait Evolution in Two Clades of New World Sparrows (Aimophila and Peucaea, Passerellidae)
Behavioral, morphological, and ecological trait evolution in two clades of New World Sparrows (Aimophila and Peucaea, Passerellidae) Carla Cicero1, Nicholas A. Mason1,4, Lauryn Benedict2 and James D. Rising3,† 1 Museum of Vertebrate Zoology, University of California, Berkeley, Berkeley, CA, United States of America 2 School of Biological Sciences, University of Northern Colorado, Greeley, CO, United States of America 3 Department of Ecology and Evolutionary Biology, University of Toronto, Toronto, Ontario, Canada 4 Current affiliation: Museum of Natural Science and Department of Biological Sciences, Louisiana State University, Baton Rouge, Louisiana, United States of America † Deceased. ABSTRACT The New World sparrows (Passerellidae) are a large, diverse group of songbirds that vary in morphology, behavior, and ecology. Thus, they are excellent for studying trait evolution in a phylogenetic framework. We examined lability versus conservatism in morphological and behavioral traits in two related clades of sparrows (Aimophila, Peucaea), and assessed whether habitat has played an important role in trait evolution. We first inferred a multi-locus phylogeny which we used to reconstruct ancestral states, and then quantified phylogenetic signal among morphological and behavioral traits in these clades and in New World sparrows more broadly. Behavioral traits have a stronger phylogenetic signal than morphological traits. Specifically, vocal duets and song structure are the most highly conserved traits, and nesting behavior appears to be maintained within clades. Furthermore, we found a strong correlation between open habitat and unpatterned plumage, complex song, and ground nesting. However, even within lineages that share the same habitat type, species vary in nesting, plumage pattern, song complexity, and duetting. Our findings highlight trade-offs between behavior, morphology, and ecology in sparrow diversification. -

Vocal Behaviour of White-Eared Ground-Sparrows (Melozone Leucotis) During the Breeding Season: Repertoires, Diel Variation, Beha
Author's personal copy J Ornithol DOI 10.1007/s10336-015-1237-y ORIGINAL ARTICLE Vocal behaviour of White-eared Ground-sparrows (Melozone leucotis) during the breeding season: repertoires, diel variation, behavioural contexts, and individual distinctiveness 1,2 2 1 Luis Sandoval • Carolina Me´ndez • Daniel J. Mennill Received: 7 March 2014 / Revised: 9 October 2014 / Accepted: 13 May 2015 Ó Dt. Ornithologen-Gesellschaft e.V. 2015 Abstract There are relatively few quantitative descriptive variation in each category of vocalisation, and found that the studies of the vocalisations and vocal behaviour of tropical Ground-sparrows produced all three vocalisations at higher bird species, in spite of the tropic’s rich avian biodiversity output at dawn (between 0500 and 0600 hours) compared to and the extensive variety of vocalisations produced by the rest of the day. This study allowed us to conduct the first tropical birds. This lack of information inhibits our under- comparisons of vocalisations between White-eared Ground- standing of tropical animals, including our ability to perform sparrows and North American species in the genus Melo- comparative analyses on vocal behaviours from an evolu- zone, and revealed both similarities and differences between tionary perspective. In this study, we present the first quan- the species groups. Our investigation also showed that titative description of the vocal repertoire and daily vocal vocalisations related to communication within pairs and to activity of White-eared Ground-sparrows (Melozone leuco- territory defence (calls and duets) exhibited lower levels of tis), using focal and autonomous recordings collected during individual distinctiveness than vocalisations related mainly two consecutive breeding seasons in Costa Rica. -

Analysis of Plumage, Morphology, and Voice Reveals Species-Level
Zootaxa 3895 (1): 103–116 ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2014 Magnolia Press ISSN 1175-5334 (online edition) http://dx.doi.org/10.11646/zootaxa.3895.1.6 http://zoobank.org/urn:lsid:zoobank.org:pub:DF4F2353-158E-4788-AD0E-73DC6F87A4D9 Analysis of plumage, morphology, and voice reveals species-level differences between two subspecies of Prevost’s Ground-sparrow Melozone biarcuata (Prévost and Des Murs) (Aves: Emberizidae) LUIS SANDOVAL1,2,3, PIERRE-PAUL BITTON1, STÉPHANIE M. DOUCET1 & DANIEL J. MENNILL1 1Department of Biological Sciences, University of Windsor, Windsor, Ontario N9B 3P4, Canada 2Escuela de Biología, Universidad de Costa Rica, San Pedro, San José, Costa Rica CP-11501-2060 3Corresponding author. E-mail: [email protected] Abstract Melozone biarcuata (Prevost’s Ground-sparrow) has traditionally been divided into two allopatric groups based on differ- ences in vocalizations and plumage characteristics: M. b. cabanisi in Costa Rica and M. b. biarcuata/M. b. hartwegi in northern Central America. However, the relationship between these subspecies has not been studied using a modern tax- onomic approach. In this study, our objective was to provide the first detailed taxonomic comparison between these three subspecies using an integrative multi-trait analysis. We analyzed morphometric features, qualitative plumage patterns, and quantitative plumage measurements using spectral reflectance from all three subspecies, and we analyzed vocalizations for subspecies M. b. biarcuata and M. b. cabanisi. Our results show that M. b. cabanisi can be readily distinguished from the two other subspecies on the basis of morphometrics (M. b. cabanisi are smaller), plumage patterns (M. -

Federal Register/Vol. 78, No. 213/Monday
65938 Federal Register / Vol. 78, No. 213 / Monday, November 4, 2013 / Proposed Rules and draft EA under Docket No. FWS– also constitutes our 12-month finding Executive Summary R6–ES–2011–0111. on a petition to remove the Inyo Purpose of Regulatory Action For additional details on specific California towhee from the Federal List information we are requesting during of Endangered and Threatened Wildlife. In 2011, we received a petition from this public comment period, please see We are seeking information and The Pacific Legal Foundation to remove the Public Comments section in our comments from the public on this from the Federal List of Endangered and September 19, 2013, Federal Register proposed rule and the post-delisting Threatened Wildlife (delist) the Inyo document (78 FR 57604), which monitoring plan. The Inyo California California towhee based on the analysis reopened the previous comment period. towhee occurs only in Inyo County, and recommendations contained in our California. 2008 5-year status review of the species Authors (Service 2008, p. 20). In 2012, we The primary authors of this document DATES: The finding announced in this published a 90-day finding (77 FR are the staff members of the Regional document was made on November 4, 32922) that concluded that the petition Office and Western Colorado Field 2013. We will accept comments presented substantial scientific or Office, Mountain-Prairie Region, U.S. received or postmarked on or before commercial information indicating that Fish and Wildlife Service. January 3, 2014. Please note that if you the petitioned action may be warranted are using the Federal eRulemaking and initiated a status review. -
Abert's Towhee Inhabits Dense Brush and Woodlands Along Sonoran Desert Rivers Diet and Foraging and Streams in Arizona and Surrounding States
Access provided by US Forest Service | In collaboration with the American Ornithological Society Explore Taxonomy Species or family ORDER Passeriformes FAMILY Passerellidae GENUS Melozone White-throated Towhee California Towhee Abert's Towhee +2 Melozone aberti LC Least Concern Names (11) Subspecies (3) WATCH Robert C. Tweit and D. M. Finch Version: 1.0 — Published March 4, 2020 Text last updated January 1, 1994 © Patrick Maurice Macaulay Library eBird LISTEN Introduction Recent analysis of mitochondrial genes suggests that the predominantly Appearance brown towhees—California, Abert's, Canyon, and White-throated—are more Systematics closely related to the Melozone ground-sparrows than they are to the predominantly black or green towhees. As a result, “brown towhees” are Distribution now placed in the genus Melozone. See the 51st Supplement to the AOU Habitat Checklist of North American Birds for details. Future revisions of this account Movements and will reflect these changes. Migration Abert's Towhee inhabits dense brush and woodlands along Sonoran Desert rivers Diet and Foraging and streams in Arizona and surrounding states. Spencer Baird described this Sounds and Vocal Behavior species in 1852 (American Ornithologists' Union 1983) and named it for Lt. James William Abert, U.S. Army (1820–1897), who obtained the specimen as a Behavior result of a survey of New Mexico at the end of the Mexican War. Abert, a West Year-round Breeding Enlarge Breeding Point graduate, served in the Topographical Engineers and retired from the Army Migration Non-Breeding Demography and after the Civil War with the rank of Lt. Colonel (Mearns and Mearns 1992a). Figure 1. -

Mapping Evaporative Water Loss in Desert Passerines Reveals an Expanding Threat of Lethal Dehydration
Mapping evaporative water loss in desert passerines reveals an expanding threat of lethal dehydration Thomas P. Albrighta,b,1, Denis Mutiibwaa,c, Alexander. R. Gersond,e, Eric Krabbe Smithe, William A. Talbote, Jacqueline J. O’Neille, Andrew E. McKechnief, and Blair O. Wolfe,1 aDepartment of Geography, University of Nevada, Reno, NV 89557; bProgram in Ecology, Evolution, and Conservation Biology, University of Nevada, Reno, NV 89557; cBeltsville Agricultural Research Center, US Department of Agriculture, Beltsville Agricultural Research Center–West, Beltsville, MD 20705; dDepartment of Biology, University of Massachusetts, Amherst, MA 01003; eBiology Department, University of New Mexico, Albuquerque, NM 87131-0001; and fDST-NRF Centre of Excellence at the Percy FitzPatrick Institute, Department of Zoology and Entomology, University of Pretoria, Hatfield 0028, South Africa Edited by Scott R. McWilliams, University of Rhode Island, Kingston, RI, and accepted by Editorial Board Member Gregory P. Asner December 21, 2016 (received for review August 16, 2016) Extreme high environmental temperatures produce a variety of maintenance of adequate hydration under conditions requiring consequences for wildlife, including mass die-offs. Heat waves are evaporative cooling (dehydration), and (ii) limits to rates of heat increasing in frequency, intensity, and extent, and are projected to dissipation and the maintenance of body temperature (Tb)below increase further under climate change. However, the spatial and lethal limits (hyperthermia). When environmental temperatures temporal dynamics of die-off risk are poorly understood. Here, we exceed a bird’s normothermic Tb, evaporative heat loss becomes the examine the effects of heat waves on evaporative water loss (EWL) only avenue of heat dissipation available to offset increasing envi- and survival in five desert passerine birds across the southwestern ronmental and internal heat loads. -

2019 ANNUAL MEETING CSUN: SCHEDULE at a GLANCE Main Events: CSU Northridge, University Student Union (USU) East
2019 ANNUAL MEETING CSUN: SCHEDULE AT A GLANCE Main Events: CSU Northridge, University Student Union (USU) East FRIDAY May 4, 2018 Symposium Symposium Time Tujunga Room Altadena Room 8:20-9:40 Desert Sciences *Rocky Reefs 9:40-9:55 Break 9:55-10:55 Desert Sciences Rocky Reefs 10:55-11:15 Break SCAS President’s Address & Awards Presentation 11:15-11:45 Location: Lake View Terrace Room Plenary Speaker: Dr. Don Prothero 11:45-12:45 Reality Check: How Science Deniers Threaten Our Future Location: Lake View Terrace Room 12:45-2:00 Lunch Symposium Contributed Papers Contributed Papers Contributed Papers Time Tujunga Room Altadena Room Van Nuys Room Panorama City Ecosystems, Habitats & 2:00-3:15 Terrestrial Ecology Parasitology Physiology Climate Change 3:15-3:35 Break Ecosystems, Habitats & 3:35-5:05 Terrestrial Ecology Parasitology Community Ecology Climate Change Poster Session 5:15-7:30 Location: Orange Grove Bistro *Rocky Reefs Symposium starts at 8:40 AM 1 Friday, May 3, 2019 Plenary Speaker REALITY CHECK: HOW SCIENCE DENIERS THREATEN OUR FUTURE Professor Donald R. Prothero, Ph.D.* Modern civilization and even human survival would not be possible without significant advances in science and medicine, yet even in the most developed countries there are people who deny the evidence of science when it conflicts with their religious or political agendas. Modern science deniers often have the same psychological factors and motivations, and typically employ the same tactics to deny reality, often using "the Holocaust Denier's playbook". In this lecture, we will examine some of the most serious forms of science denialism, from climate deniers to creationists to anti-vaxxers, why they resist the discoveries of science yet embrace the latest technology and medicine, and what it means for our future.