(SSC) Region of Chloroplast Genomes1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Origin of Alternation of Generations in Land Plants
Theoriginof alternation of generations inlandplants: afocuson matrotrophy andhexose transport Linda K.E.Graham and LeeW .Wilcox Department of Botany,University of Wisconsin, 430Lincoln Drive, Madison,WI 53706, USA (lkgraham@facsta¡.wisc .edu ) Alifehistory involving alternation of two developmentally associated, multicellular generations (sporophyteand gametophyte) is anautapomorphy of embryophytes (bryophytes + vascularplants) . Microfossil dataindicate that Mid ^Late Ordovicianland plants possessed such alifecycle, and that the originof alternationof generationspreceded this date.Molecular phylogenetic data unambiguously relate charophyceangreen algae to the ancestryof monophyletic embryophytes, and identify bryophytes as early-divergentland plants. Comparison of reproduction in charophyceans and bryophytes suggests that the followingstages occurredduring evolutionary origin of embryophytic alternation of generations: (i) originof oogamy;(ii) retention ofeggsand zygotes on the parentalthallus; (iii) originof matrotrophy (regulatedtransfer ofnutritional and morphogenetic solutes fromparental cells tothe nextgeneration); (iv)origin of a multicellularsporophyte generation ;and(v) origin of non-£ agellate, walled spores. Oogamy,egg/zygoteretention andmatrotrophy characterize at least some moderncharophyceans, and arepostulated to represent pre-adaptativefeatures inherited byembryophytes from ancestral charophyceans.Matrotrophy is hypothesizedto have preceded originof the multicellularsporophytes of plants,and to represent acritical innovation.Molecular -

Cell Wall Ribosomes Nucleus Chloroplast Cytoplasm
Cell Wall Ribosomes Nucleus Nickname: Protector Nickname: Protein Maker Nickname: Brain The cell wall is the outer covering of a Plant cell. It is Ribosomes read the recipe from the The nucleus is the largest organelle in a cell. The a strong and stiff and made of DNA and use this recipe to make nucleus directs all activity in the cell. It also controls cellulose. It supports and protects the plant cell by proteins. The nucleus tells the the growth and reproduction of the cell. holding it upright. It ribosomes which proteins to make. In humans, the nucleus contains 46 chromosomes allows water, oxygen and carbon dioxide to pass in out They are found in both plant and which are the instructions for all the activities in your of plant cell. animal cells. In a cell they can be found cell and body. floating around in the cytoplasm or attached to the endoplasmic reticulum. Chloroplast Cytoplasm Endoplasmic Reticulum Nickname: Oven Nickname: Gel Nickname: Highway Chloroplasts are oval structures that that contain a green Cytoplasm is the gel like fluid inside a The endoplasmic reticulum (ER) is the transportation pigment called chlorophyll. This allows plants to make cell. The organelles are floating around in center for the cell. The ER is like the conveyor belt, you their own food through the process of photosynthesis. this fluid. would see at a supermarket, except instead of moving your groceries it moves proteins from one part of the cell Chloroplasts are necessary for photosynthesis, the food to another. The Endoplasmic Reticulum looks like a making process, to occur. -

Brown Algae and 4) the Oomycetes (Water Molds)
Protista Classification Excavata The kingdom Protista (in the five kingdom system) contains mostly unicellular eukaryotes. This taxonomic grouping is polyphyletic and based only Alveolates on cellular structure and life styles not on any molecular evidence. Using molecular biology and detailed comparison of cell structure, scientists are now beginning to see evolutionary SAR Stramenopila history in the protists. The ongoing changes in the protest phylogeny are rapidly changing with each new piece of evidence. The following classification suggests 4 “supergroups” within the Rhizaria original Protista kingdom and the taxonomy is still being worked out. This lab is looking at one current hypothesis shown on the right. Some of the organisms are grouped together because Archaeplastida of very strong support and others are controversial. It is important to focus on the characteristics of each clade which explains why they are grouped together. This lab will only look at the groups that Amoebozoans were once included in the Protista kingdom and the other groups (higher plants, fungi, and animals) will be Unikonta examined in future labs. Opisthokonts Protista Classification Excavata Starting with the four “Supergroups”, we will divide the rest into different levels called clades. A Clade is defined as a group of Alveolates biological taxa (as species) that includes all descendants of one common ancestor. Too simplify this process, we have included a cladogram we will be using throughout the SAR Stramenopila course. We will divide or expand parts of the cladogram to emphasize evolutionary relationships. For the protists, we will divide Rhizaria the supergroups into smaller clades assigning them artificial numbers (clade1, clade2, clade3) to establish a grouping at a specific level. -

The Structure and Function of Plastids
The Structure and Function of Plastids Edited by Robert R. Wise University of Wisconsin, Oshkosh WI, USA and J. Kenneth Hoober Arizona State University, Tempe AZ, USA Contents From the Series Editor v Contents xi Preface xix A Dedication to Pioneers of Research on Chloroplast Structure xxi Color Plates xxxiii Section I Plastid Origin and Development 1 The Diversity of Plastid Form and Function 3–25 Robert R. Wise Summary 3 I. Introduction 4 II. The Plastid Family 5 III. Chloroplasts and their Specializations 13 IV. Concluding Remarks 20 Acknowledgements 21 References 21 2 Chloroplast Development: Whence and Whither 27–51 J. Kenneth Hoober Summary 27 I. Introduction 28 II. Brief Review of Plastid Evolution 28 III. Development of the Chloroplast 32 IV. Overview of Photosynthesis 43 References 46 3 Protein Import Into Chloroplasts: Who, When, and How? 53–74 Ute C. Vothknecht and J¨urgen Soll Summary 53 I. Introduction 54 II. On the Road to the Chloroplast 56 III. Protein Translocation via Toc and Tic 58 IV. Variations on Toc and Tic Translocation 63 V. Protein Translocation and Chloroplast Biogenesis 64 VI. The Evolutionary Origin of Toc and Tic 66 VII. Intraplastidal Transport 66 VIII. Protein Translocation into Complex Plastids 69 References 70 xi 4 Origin and Evolution of Plastids: Genomic View on the Unification and Diversity of Plastids 75–102 Naoki Sato Summary 76 I. Introduction: Unification and Diversity 76 II. Endosymbiotic Origin of Plastids: The Major Unifying Principle 78 III. Origin and Evolution of Plastid Diversity 85 IV. Conclusion: Opposing Principles in the Evolution of Plastids 97 Acknowledgements 98 References 98 5 The Mechanism of Plastid Division: The Structure and Origin of The Plastid Division Apparatus 103–121 Shin-ya Miyagishima and Tsuneyoshi Kuroiwa Summary 104 I. -

Algal Chloroplasts Secondary Article
Algal Chloroplasts Secondary article Saul Purton, University College London, London, UK Article Contents . Introduction A great diversity of chloroplasts is found amongst the various algal groups. This diversity . Diversity and Classification of Algae is the result of an intriguing evolutionary process that involved the acquisition of . Origins and Evolution of Algal Chloroplasts chloroplasts by different eukaryotic organisms. Chloroplast Genetics and Molecular Biology . Protein Transport in the Chloroplast . Summary Introduction The chloroplast is one of a family of related biosynthetic and which lack the differentiated structures that organelles (termed plastids) found within the cells of define higher plants (roots, shoots, leaves, etc.). Indeed, plants, eukaryotic algae and certain protists. The primary the algae are often referred to as ‘lower’ or ‘primitive’ role of the chloroplast is the fixation of atmospheric carbon plants. Included within the algae are the prokaryotic through photosynthesis, but it is also the site of synthesis of cyanobacteria (formerly referred to as blue-green algae), many other important compounds including pigments, together with a diverse collection of microscopic and fatty acids, amino acids and nucleotides. Chloroplasts are macroscopic eukaryotes. Algal species can be unicellular, distinguishable from other plastid types in that they filamentous or multicellular and they range in size from the contain chlorophyll and other pigments that are involved unicellular forms that are only a few micrometres in in light energy capture and dissipation. In higher plants, diameter to the giant Laminaria seaweeds that are tens of nonphotosynthetic plastids such as chromoplasts, amylo- metres long. Algae have adapted to life in a wide range of plasts and leucoplasts are found in nongreen tissue and environments. -

Chloroplasts Are the Food Producers of the Cell. the Organelles Are Only Found in Plant Cells and Some Protists Such As Algae
Name: ___________________________ Cell #2 H.W. due September 22nd, 2016 Period: _________ Chloroplasts are the food producers of the cell. The organelles are only found in plant cells and some protists such as algae. Animal cells do not have chloroplasts. Chloroplasts work to convert light energy of the Sun into sugars that can be used by cells. It is like a solar panel that changes sunlight energy into electric energy. The entire process is called photosynthesis and it all depends on the little green chlorophyll molecules in each chloroplast. In the process of photosynthesis, plants create sugars and release oxygen (O2). The oxygen released by the chloroplasts is the same oxygen you breathe every day. Chloroplasts are found in plant cells, but not in animal cells. The purpose of the chloroplast is to make sugars that feed the cell’s machinery. Photosynthesis is the process of a plant taking energy from the Sun and creating sugars. When the energy from the Sun hits a chloroplast and the chlorophyll molecules, light energy is converted into the chemical energy. Plants use water, carbon dioxide, and sunlight to make sugar and oxygen. During photosynthesis radiant energy or solar energy or light energy is transferred into chemical energy in the form of sugar (glucose). You already know that during photosynthesis plants make their own food. The food that the plant makes is in the form of sugar that is used to provide energy for the plant. The extra sugar that the plant does not use is stored as starch for later use. Mitochondria are known as the powerhouses of the cell. -

Chloroplast Genes Are Expressed During Intracellular Symbiotic
Proc. Natl. Acad. Sci. USA Vol. 93, pp. 12333-12338, October 1996 Cell Biology Chloroplast genes are expressed during intracellular symbiotic association of Vaucheria litorea plastids with the sea slug Elysia chlorotica (photosystem II reaction center/photosynthesis/chromophytic alga/ascoglossan mollusc/gene expression) CESAR V. MUJER*t, DAVID L. ANDREWS*t, JAMES R. MANHART§, SIDNEY K. PIERCES, AND MARY E. RUMPHO*II Departments of *Horticultural Sciences and §Biology, Texas A & M University, College Station, TX 77843; and IDepartment of Zoology, University of Maryland, College Park, MD 20742 Communicated by Martin Gibbs, Brandeis University, Waltham, MA, August 16, 1996 (received for review January 26, 1996) ABSTRACT The marine slug Elysia chlorotica (Gould) lowing metamorphosis from the veliger stage when juvenile forms an intracellular symbiosis with photosynthetically ac- sea slugs begin to feed on V litorea cells (1, 2). Once ingested, tive chloroplasts from the chromophytic alga Vaucheria litorea the chloroplasts are phagocytically incorporated into the cy- (C. Agardh). This symbiotic association was characterized toplasm of one of two morphologically distinct, epithelial cells over a period of 8 months during which E. chlorotica was (3) and maintain their photosynthetic function (1, 3). The deprived of V. litorea but provided with light and CO2. The fine plastids are frequently found in direct contact with the host structure of the symbiotic chloroplasts remained intact in E. cytoplasm as revealed by ultrastructural studies (3). In nature, chlorotica even after 8 months of starvation as revealed by the adult animal feeds on algae only sporadically, obtaining electron microscopy. Southern blot analysis of total DNA metabolic energy from the photosynthetic activity of the from E. -

Arabidopsis EGY1 Is Critical for Chloroplast Development in Leaf Epidermal Guard Cells
plants Article Arabidopsis EGY1 Is Critical for Chloroplast Development in Leaf Epidermal Guard Cells Alvin Sanjaya 1,†, Ryohsuke Muramatsu 1,†, Shiho Sato 1, Mao Suzuki 1, Shun Sasaki 1, Hiroki Ishikawa 1, Yuki Fujii 1, Makoto Asano 1, Ryuuichi D. Itoh 2, Kengo Kanamaru 3, Sumie Ohbu 4, Tomoko Abe 4, Yusuke Kazama 4,5 and Makoto T. Fujiwara 1,4,* 1 Department of Materials and Life Sciences, Faculty of Science and Technology, Sophia University, Kioicho, Chiyoda, Tokyo 102-8554, Japan; [email protected] (A.S.); [email protected] (R.M.); [email protected] (S.S.); [email protected] (M.S.); [email protected] (S.S.); [email protected] (H.I.); [email protected] (Y.F.); [email protected] (M.A.) 2 Department of Chemistry, Biology and Marine Science, Faculty of Science, University of the Ryukyus, Okinawa 903-0213, Japan; [email protected] 3 Faculty of Agriculture, Kobe University, Nada, Kobe 657-8501, Japan; [email protected] 4 RIKEN Nishina Center, Wako, Saitama 351-0198, Japan; [email protected] (S.O.); [email protected] (T.A.); [email protected] (Y.K.) 5 Faculty of Bioscience and Biotechnology, Fukui Prefectural University, Eiheiji, Fukui 910-1195, Japan * Correspondence: [email protected]; Tel.: +81-3-3238-3495 † These authors contributed equally to this work. Abstract: In Arabidopsis thaliana, the Ethylene-dependent Gravitropism-deficient and Yellow-green 1 (EGY1) gene encodes a thylakoid membrane-localized protease involved in chloroplast development in leaf Citation: Sanjaya, A.; Muramatsu, R.; mesophyll cells. -
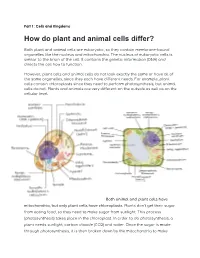
How Do Plant and Animal Cells Differ?
Part 1: Cells and Kingdoms How do plant and animal cells differ? Both plant and animal cells are eukaryotic, so they contain membrane-bound organelles like the nucleus and mitochondria. The nucleus of eukaryotic cells is similar to the brain of the cell. It contains the genetic information (DNA) and directs the cell how to function. However, plant cells and animal cells do not look exactly the same or have all of the same organelles, since they each have different needs. For example, plant cells contain chloroplasts since they need to perform photosynthesis, but animal cells do not. Plants and animals are very different on the outside as well as on the cellular level. Both animal and plant cells have mitochondria, but only plant cells have chloroplasts. Plants don’t get their sugar from eating food, so they need to make sugar from sunlight. This process (photosynthesis) takes place in the chloroplast. In order to do photosynthesis, a plant needs sunlight, carbon dioxide (CO2) and water. Once the sugar is made through photosynthesis, it is then broken down by the mitochondria to make energy for the cell. Because animals get sugar from the food they eat, they do not need chloroplasts: just mitochondria. Both plant and animal cells have vacuoles. A plant cell contains a large, singular vacuole that is used for storage of water and nutrients. It also helps maintain the shape of the cell. In contrast, animal cells have many, smaller vacuoles, which also are used for storage of water and nutrients. Plant cells have a cell wall, as well as a cell membrane. -

Phylogenomic Insights Into the Origin of Primary Plastids
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.03.231043; this version posted August 4, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Phylogenomic Insights into the Origin of Primary Plastids 2 3 Iker Irisarri1,2,3,*, Jürgen F. H. Strassert1,4, Fabien Burki1,5 4 5 1 Department of Organismal Biology (Systematic Biology), Uppsala University, Norbyv. 6 18D, 75236 Uppsala, Sweden 7 2 Department of Biodiversity and Evolutionary Biology, Museo Nacional de Ciencias 8 Naturales, José Gutiérrez Abascal 2, 28006 Madrid, Spain 9 3 Current address: Department of Applied Bioinformatics, Institute for Microbiology 10 and Genetics, University of Göttingen, Goldschmidtstr. 1, Göttingen, Germany 11 4 Department of Biology, Chemistry, and Pharmacy, Institute of Biology, Evolutionary 12 Biology, Free University of Berlin, Königin-Luise-Str. 1–3, 14195 Berlin, Germany 13 5 Science For Life Laboratory, Uppsala University, 75236 Sweden 14 15 * Email: [email protected] 16 bioRxiv preprint doi: https://doi.org/10.1101/2020.08.03.231043; this version posted August 4, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 17 Abstract 18 19 The origin of plastids was a major evolutionary event that paved the way for an 20 astonishing diversification of photosynthetic eukaryotes. -

Chloroplasts in Living Cells and the String-Of-Grana Concept of Chloroplast Structure Revisited
UCLA UCLA Previously Published Works Title Chloroplasts in living cells and the string-of-grana concept of chloroplast structure revisited Permalink https://escholarship.org/uc/item/5d448880 Journal Photosynthesis Research, 80(1-3) ISSN 0166-8595 Authors Wildman, S G Hirsch, Ann M Kirchanski, S J et al. Publication Date 2004 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Photosynthesis Research 077G: 1–8, 2003. 1 © 2003 Kluwer Academic Publishers. Printed in the Netherlands. Perspective Chloroplasts in living cells and the string-of-grana concept of chloroplast structure revisited S. G. Wildman1,AnnM.Hirsch1,2,∗,S.J.Kirchanski3 & Donald Spencer4 1Department of Molecular, Cell and Developmental Biology, University of California-Los Angeles, Los Angeles, CA 90095-1606, USA; 2Molecular Biology Institute, UCLA, Los Angeles, CA 90095-1606, USA; 3ReedSmith, 1900 Avenue of the Stars, Los Angeles, CA 90067, USA; 4CSIRO Plant Industry, GPO Box 1600, Canberra, Australia 2601; ∗Author for correspondence (e-mail: [email protected]; fax: +1-310-206-5413) Received 14 November 2002; accepted in revised form 14 May 2003 Key words: grana, living chloroplasts, string-of-grana model Abstract In 1980, Wildman et al. proposed a three-dimensional model for chloroplast structure whereby the grana were arranged in non-overlapping rows, like beads on a string. This string-of-grana model was developed from phase microscope analysis of living cells and partially disrupted, isolated chloroplasts. However, models based on anal- yses by various electron microscope (EM) techniques (which inevitably encompass a relatively small fraction of the whole chloroplast) indicated that grana are interconnected in all directions by intergranal lamellae and not just along a single ‘string’. -

Photosynthesis
Photosynthesis Photosynthesis is the process by which plants, some bacteria and some protistans use the energy from sunlight to produce glucose from carbon dioxide and water. This glucose can be converted into pyruvate which releases adenosine triphosphate (ATP) by cellular respiration. Oxygen is also formed. Photosynthesis may be summarised by the word equation: carbon dioxide + water glucose + oxygen The conversion of usable sunlight energy into chemical energy is associated with the action of the green pigment chlorophyll. Chlorophyll is a complex molecule. Several modifications of chlorophyll occur among plants and other photosynthetic organisms. All photosynthetic organisms have chlorophyll a. Accessory pigments absorb energy that chlorophyll a does not absorb. Accessory pigments include chlorophyll b (also c, d, and e in algae and protistans), xanthophylls, and carotenoids (such as beta-carotene). Chlorophyll a absorbs its energy from the violet-blue and reddish orange-red wavelengths, and little from the intermediate (green-yellow-orange) wavelengths. Chlorophyll All chlorophylls have: • a lipid-soluble hydrocarbon tail (C20H39 -) • a flat hydrophilic head with a magnesium ion at its centre; different chlorophylls have different side-groups on the head The tail and head are linked by an ester bond. Leaves and leaf structure Plants are the only photosynthetic organisms to have leaves (and not all plants have leaves). A leaf may be viewed as a solar collector crammed full of photosynthetic cells. The raw materials of photosynthesis, water and carbon dioxide, enter the cells of the leaf, and the products of photosynthesis, sugar and oxygen, leave the leaf. Water enters the root and is transported up to the leaves through specialized plant cells known as xylem vessels.