Book of Abstracts
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Studies on Variability, Heritability and Genetic Advance in Double Type
International Journal of Chemical Studies 2020; 8(1): 3101-3104 P-ISSN: 2349–8528 E-ISSN: 2321–4902 www.chemijournal.com Studies on variability, heritability and genetic IJCS 2020; 8(1): 3101-3104 © 2020 IJCS advance in double type genotypes of tuberose Received: 26-11-2019 Accepted: 28-12-2019 (Agave amica (Medik.) Syn. Polianthes tuberosa L.) Ratna Priyanka R Ph.D. Scholar, Department of Floriculture and Landscaping, Tamil Nadu Agricultural Ratna Priyanka R, M Kannan, M Ganga and N Kumaravadivel University, Coimbatore, Tamil Nadu, India DOI: https://doi.org/10.22271/chemi.2020.v8.i1au.8741 M Kannan Professor, Directorate of Abstract Research, Tamil Nadu The present study on performance and genetic variability studies in tuberose (Agave amica (Medik.) Syn. Agricultural University, Polianthes tuberosa L.) Double type genotypes was conducted at Botanical Garden, Department of Coimbatore, Tamil Nadu, India Floriculture and Landscape Architecture, Tamil Nadu Agricultural University during the year 2018-2020. Among five double type genotypes, Suvasini followed by Hyderabad Double and Vaibhav showed better M Ganga performance in vegetative, flowering and yield parameters with spike yield of 3.38, 3.31 and 3.25 spikes/ Associate Professor (Hort.), plant respectively and bulb yield of 735.00, 654.75 and 540.75 g/ plant respectively. Among different Department of Floriculture and vegetative and flowering parameters recorded, phenotypic coefficient of variation and genotypic Landscape Architecture, Tamil coefficient of variation were found to -

PLANT NAME COMMON NAME ZONES DESCRIPTION # PLANTS LOCA Abutilons Vines PLANT NAME COMMON NAME ZONES DESCRIPTION # PLANTS LOCA Ta
COMMON # PLANT NAME NAME ZONES DESCRIPTION PLANTS LOCA LOCA = Locations R = Row, Tbl = Table, GH1 = Green House 1, GH2 = Green House 2 Evergreen perennials with upright, arching growth from from 10" to 8' depending on variety. Main bloom season in spring, but can bloom all year. Abutilons Flowering Maple 12-24 Dwarf Red Really red-orange, 15"-18" Row 16 Low, compact blossoms are up to 2" across, pale Halo apricot, 4'-5', ok down to 12-15o F with protection. 28 Tbl 1 Peach peach blossoms, 6'-8'. Greyish green leaves with broad ivory margins and Savitzii apricot pink flowers. 3'-4' 33 GH2, R16 Broad green leaves edged in creamy white with pink Souvenir de Bonn flowers. 3'-9' R16 Tangerine 6'-8' , tangerine orange blossoms R16 Drooping red and yellow blossoms, 4'-8'. Attracts Teardrop butterflies and hummingbirds. R16 megapotamicum Bright speckled foliage, somewhat vining, 3'-5'. "Paisley" Appreciates some pruning, attracts hummingbirds. 5 Shade 6' H & W, Salmon orange blossoms with broad green Victor Reiter leaves. 3 R 16 Vines 15-20', fast growing, vigorous vine somewhat frost tender, beautiful coral flowers. Best in full sun or part- shade. Moderate water, does not tolerate heat well. Passiflora Coral Seas 19-24 Needs frost protection. 48 GH2 12', modest climber, bears 4" blue flowers with yellow center, fragrant. Full sun or part shade, moderate water. Poisonous if ingested. Solanum crispum Chilean Potato Vine 12-24 18 Tbl 1 COMMON # PLANT NAME NAME ZONES DESCRIPTION PLANTS LOCA Tall Perennials Lemon scented foliage with pink flowers in dense flower heads. -
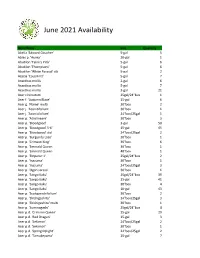
June 2021 Availability
June 2021 Availability Item Name Size Quantity Abelia 'Edward Goucher' 5-gal 5 Abies p. 'Aurea' 20-gal 1 Abutilon 'Talini's Pink' 5-gal 6 Abutilon 'Thompsonii' 5-gal 6 Abutilon 'White Parasol' stk 5-gal 2 Acacia 'Cousin Itt' 5-gal 7 Acanthus mollis 2-gal 6 Acanthus mollis 5-gal 7 Acanthus mollis 3-gal 21 Acer circinatum 25gal/24"box 1 Acer f. 'Autumn Blaze' 15-gal 6 Acer g. 'Flame' multi 30"box 2 Acer j. 'Aconitifolium' 30"box 1 Acer j. 'Aconitifolium' 24"box/25gal 1 Acer p. 'Atrolineare' 30"box 3 Acer p. 'Bloodgood' 3-gal 50 Acer p. 'Bloodgood' 5-6' 15-gal 45 Acer p. 'Bloodgood' std. 24"box/25gal 2 Acer p. 'Burgundy Lace' 30"box 1 Acer p. 'Crimson King' 36"box 6 Acer p. 'Emerald Queen 36"box 1 Acer p. 'Emerald Queen 48"box 1 Acer p. 'Emperor 1' 25gal/24"box 2 Acer p. 'Inazuma' 30"box 1 Acer p. 'Inazuma' 24"box/25gal 3 Acer p. 'Ogon sarasa' 30"box 1 Acer p. 'Sango Kaku' 25gal/24"box 39 Acer p. 'Sango Kaku' 15-gal 41 Acer p. 'Sango Kaku' 30"box 4 Acer p. 'Sango Kaku' 10-gal 43 Acer p. 'Scolopendrifolium' 30"box 2 Acer p. 'Shishigashira' 24"box/25gal 3 Acer p. 'Shishigashira' multi 30"box 1 Acer p. 'Suminagashi' 25gal/24"box 4 Acer p.d. 'Crimson Queen' 15-gal 29 Acer p.d. 'Red Dragon' 15-gal 3 Acer p.d. 'Sekimori' 24"box/25gal 2 Acer p.d. 'Sekimori' 30"box 1 Acer p.d. -

Ministério Da Agricultura, Pecuária E Abastecimento
Seção 1 ISSN 1677-7042 Nº 232, sexta-feira, 4 de dezembro de 2020 S EC R E T A R I A - G E R A L I - Ministro de Estado Chefe da Secretaria-Geral da Presidência da República, que o coordenará; PORTARIA Nº 98, DE 3 DE DEZEMBRO DE 2020 II - Secretário-Executivo; Aprova o Plano de Dados Abertos da Secretaria-Geral da Presidência da República -vigência 2020-2022. III - Secretário Especial de Modernização do Estado; O MINISTRO DE ESTADO CHEFE DA SECRETARIA-GERAL DA PRESIDÊNCIA DA IV - Secretário Especial de Administração; REPÚBLICA , no uso das atribuições que lhe conferem o artigo 87 da Constituição Federal, V - Secretário de Controle Interno; e considerando o que dispõem a Lei nº 12.527, de 18 de outubro de 2011, o Decreto nº 8.777, de 11 de maio de 2016, e a Resolução CGINDA nº 03, de 13 de outubro de 2017, VI - Subchefe para Assuntos Jurídicos; e que disciplinam a Política de Dados Abertos do Poder Executivo Federal, resolve: VII - Diretor-Geral da Imprensa Nacional. Art. 1º Aprovar o Plano de Dados Abertos da Secretaria-Geral da Presidência da § 1º Os membros de que tratam os incisos do caput serão representados, em República - vigência 2020-2022. suas ausências e impedimentos por seus substitutos no cargo em comissão ou função de confiança que ocupam. Art. 2º O referido Plano torna público o inventário de bases de dados do órgão,o § 2º O Gestor de Segurança da Informação da Secretaria-Geral da Presidência da resultado da consulta pública que identificou o interesse público pelas bases da SG/PR, a seleção República participará das reuniões do Comitê que tenham como pauta o tema da segurança dos dados que serão abertos, o cronograma de publicação, a descrição de ações de fomento ao da informação, coordenando, com direito a voto, a sessão específica referente ao tema. -

Pdf 753.12 K
MBTJ, 3(3): 117-121, 2019 Research Paper Investigation of hormone effects on reproductive properties of Agave amica in different grow condition Ali Zaher*, Osman Rahmati Department of Biology, Faculty of Science, Kabul University, Afghanistan Received: 11 July 2019 Accepted: 22 August 2019 Published: 01 September 2019 Abstract Agave amica (Polianthes tuberosa L) is one of the most important cut flowers in Middle East and the world and it is widely used in flower decoration due to its beauty and perfume [1]. To investigate the effects of plant density and Gibberellin treatment on tuberose bulbs, an experiment was conducted using a Split Plot based on Randomized Complete Block Design with three replications in the Jiroft city. In this experiment, plant density in three levels (10, 15, and 25 plants per m2) as the main factor and gibberellin concentrations in four levels (control, 100, 200, and 300 ppm) as a sub-factor were considered. The present study was performed in 12 treatments and three replications, which consisted of 36 experimental plots. The results showed that the studied indices were affected by the application of gibberellin and plant density treatments, except for the number of cromels per plant. Also, the highest values in the number of leaf per plant, inflorescence length and number of floret per inflorescence indices were obtained in the mentioned treatment, and the lowest amounts of them were observed in the density of 25 plants per m2 × no application of gibberellin. Therefore, the best treatment for the cultivation of tuberose plant was the application of 300 ppm of gibberellic acid and the density of 15 plants per m2. -

Floral Intimate Credits
Floral Intimate Credits: © TERRE Policy Centre Publication: TERRE Policy Centre Presented By: Dr. Vinita Apte, Founder Director, TERRE Policy Centre Title of Book: Floral Intimate Year of Publication: 2020 Place of Publication: Pune Floral Designed by: Mr. Avinash R. Kulkarni Scientific name Crossandra infundibuliformis Common Name Firecracker flower, Aboli It is usually grown in containers but can be attractive in beds as well. The flowers have no perfume but stay fresh for several days on the bush. A well-tended specimen will bloom continuously for years. The tiny flowers are often strung together into strands, sometimes along with white jasmine flowers and therefore in great demand for making garlands which are offered to temple deities or used to embellish women's hair. Scientific Name Bougainvillea Common Name Bouganvel Bougainvillea are popular ornamental plants in most areas with warm climates. It is referred to as "paper flower" because the bracts are thin and papery. Flowers are of bright colours associated with the plant, including pink, magenta, purple, red, orange, white, or yellow. Scientific Name Saussurea obvallata Common Name Brahm Kamal. Found in the region of the Himalayas, Brahma Kamal is considered a medicinal herb in Tibetian Medicine. It is used to treat urogenital disorders, liver infections, sexually transmitted diseases, bone pains, and cold and cough. It has a bitter taste and the entire plant is used. The plant is endangered because people cut it down for their own use. Scientific Name Scadoxus multiflorus Common Name Ball Lily, Blood Lily It is a bulbous plant native to most of sub –Saharan Africa from Senegal to Soma lia to South Africa. -

Value Addition of Loose Flowers
© 2021 JETIR May 2021, Volume 8, Issue 5 www.jetir.org (ISSN-2349-5162) Value Addition of Loose Flowers 1Oyem Kombo, 2Dr. Homraj Anandrao Sahare 1 MSc. Student, 2Assistant Professor 1Department of floriculture and landscaping, Lovely Professional University, Phagwara, Punjab, India. Abstract: Value addition is the process in which for the same volume of a primary product, a high price is obtained by means of processing, packaging, upgrading the quality and various other methods. Value addition in floriculture is a process to improve the economic value and also employment. The value addition of in floriculture includes adopting proper post harvest technology and logistics. The value addition helps to reduce the post harvest loss and to provide better quality, empower farmers and other weaker sections. There are numbers of flowers having potential value addition venture. Index terms: Loose flower, value addition, shelf life, essential oil, absolute. 1. Introduction: Floriculture industry is presently considered as one of the most profit making sector in agro-enterprise. Flowers have various uses in human day today life due to its aesthetic value, varying colors, fragrance, texture, diverse form (Shweta, 2018). But flowers being highly perishable, makes it challenging to promote in market following inadequate post-harvest management practices for the growers. Floriculture sector not only includes the loose flowers, cut flowers and ornamentals but also the value added products derived from the flowers and its parts fetching really good amount of money in market and several industries increasing economic value and providing employments. Value addition of flowers is the process of improving economic value and appeal of any floriculture products by deriving changes in genetics and through processing as well (Mebakerlin et al, 2015) using innovative methods. -

Author's Blurb
Author’s Blurb TK Lim (Tong Kwee Lim) obtained his Bachelor products into and out of Australia for the Middle and Masters in Agricultural Science from the East and Asian region. During his time with University of Malaya and his PHD (Botanical ACIAR, he oversaw and managed international Sciences) from the University of Hawaii. He research and development programs in plant pro- worked in the University of Agriculture Malaysia tection and horticulture covering a wide array of for 20 years as a lecturer and Associate Professor; crops that included fruits, plantation crops, veg- as Principal Horticulturist for 9 years for the etables, culinary and medicinal herbs and spices Department of Primary Industries and Fisheries, mainly in southeast Asia and the Pacifi c. In the Darwin, Northern Territory; 6 years as Manager course of his four decades of working career he of the Asia and Middle East Team in Plant has travelled extensively worldwide to many Biosecurity Australia, Department of Agriculture, countries in South Asia, East Asia, southeast Fisheries and Forestry, Australia; and 4 years as Asia, Middle East, Europe, the Pacifi c Islands, Research Program Manager with the Australian USA and England, and also throughout Malaysia Centre for International Agriculture Research and Australia. Since his tertiary education days (ACIAR), Department of Foreign Affairs and he always had a strong passion for crops and took Trade, Australia before he retired from public an avid interest in edible and medicinal plants. service. He has published over a hundred -

Botany Part 2 at End
B O T A N Y L I S T I N G S PART 1 1970-2013 Updated 26.07.21 There are no orchids sets on this list. Please ask for our separate Orchids list PHILATELIC SUPPLIES (M.B.O'Neill) 359 Norton Way South Letchworth Garden City HERTS ENGLAND SG6 1SZ (Telephone 0044-(0)1462-684191 during office hours 9.30-3.-00pm UK time Mon.-Fri.) Web-site: www.philatelicsupplies.co.uk email: [email protected] TERMS OF BUSINESS: & Notes on these lists: (Please read before ordering). 1). All stamps are unmounted mint unless specified otherwise. we aim to be HALF-CATALOGUE PRICE OR UNDER 2). Lists are updated about every 6 months to include most recent stock movements and New Issues; they are therefore reasonably accurate stockwise & 100% pricewise. This reduces the need for "credit notes" or refunds. Alternatives may be listed in case items are out of stock However, these popular lists are still best used as soon as possible. Next listings will be printed in 6 & 12 months time; please say when next we should send a list. 3). New Issues Services can be provided if you wish to keep your collection up to date on a Standing Order basis. Details & forms on request. Regret we do not run an on approval service. 4). All orders on our order forms are attended to by return of post. We will keep a photocopy it and return your annotated original. 5). Other Thematic Lists are available on request. Orchids, Birds, etc. ; Botany Part 2 at end 6). -

Some Recommended 'Extra-Scent-Tial' Fragrance Plants for PNW Gardens
Some Recommended ‘Extra-scent-tial’ Fragrance plants for PNW Gardens Scent time Hardine Mature Size Type Exposure/Cultural Notes ss Zones (HXW) Plant Name Sp S F W Daphne Full sun to light shade Semi-evergreen Daphne x transatlantica “Summer Ice” 6-8 4’X6’ Provide good drainage; don’t disturb x x x Shrub (+ others) roots Perfumes whole garden Star Jasmine 10’ - 20’ with Trachelospermum jasminoides support; can Evergreen vine be kept shorter Perfumes whole garden 7-8 Full sun to part shade in hot areas x x x or ground cover or left to scramble Sweet Box Sarcococca orientalis, Sarcococca confusa Evergreen Deep to light shade, will take dry once Sarcococca ruscifolia 6-9 1.5’X3’ x x shrub established All perfume whole garden Sweet Bay Magnolia Early summer and Magnolia virginiana Small evergreen 6-9 20’T x 18’w Full sun to one third light shade then sporadic through ‘Moonglow’ is a good cultivar tree Lemon scented flowers and crushed season. leaves smell like bayleaf. Rhododendrons and Azaleas Extremely fragrant ones: Azalea ‘Fragrant Star’ Deciduous or Rhododendron ‘Fragrantissimum’ evergreen varies varies Varies x R. ‘Loderi King George’ shrubs Azalea ‘Weston’s Innocence’ Many others -- Gardenia.net has an excellent list. Annual vine or Full sun, keep watered and pick Sweet Pea varies x x Lathyrus odoratus bush regularly or dead head to extend bloom Deciduous Lilac Full sun. Not all are fragrant so do shrub or small 3-8 varies x Syringa species and hybrids research. Some are resistant to mildew tree Evergreen Clematis Full sun to part shade, shaded/cool Clematis armandii Vine 7-9 20’, BIG Late winter to spring roots Perfumes whole garden 8-11 Nicotiana, Jasmine Tobacco Annual to Full sun to part shade; most fragrant in possibly 2’X5’ x x x Nicotiana alata tender perennial the evening lower. -
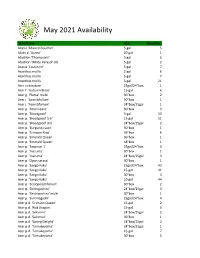
May 2021 Availability
May 2021 Availability Item Name Size Quantity Abelia 'Edward Goucher' 5-gal 5 Abies p. 'Aurea' 20-gal 1 Abutilon 'Thompsonii' 5-gal 6 Abutilon 'White Parasol' stk 5-gal 2 Acacia 'Cousin Itt' 5-gal 7 Acanthus mollis 2-gal 6 Acanthus mollis 5-gal 7 Acanthus mollis 3-gal 21 Acer circinatum 25gal/24"box 1 Acer f. 'Autumn Blaze' 15-gal 6 Acer g. 'Flame' multi 30"box 2 Acer j. 'Aconitifolium' 30"box 1 Acer j. 'Aconitifolium' 24"box/25gal 1 Acer p. 'Atrolineare' 30"box 3 Acer p. 'Bloodgood' 3-gal 50 Acer p. 'Bloodgood' 5-6' 15-gal 51 Acer p. 'Bloodgood' std. 24"box/25gal 2 Acer p. 'Burgundy Lace' 30"box 1 Acer p. 'Crimson King' 36"box 6 Acer p. 'Emerald Queen 36"box 1 Acer p. 'Emerald Queen 48"box 1 Acer p. 'Emperor 1' 25gal/24"box 3 Acer p. 'Inazuma' 30"box 1 Acer p. 'Inazuma' 24"box/25gal 3 Acer p. 'Ogon sarasa' 30"box 1 Acer p. 'Sango Kaku' 25gal/24"box 43 Acer p. 'Sango Kaku' 15-gal 41 Acer p. 'Sango Kaku' 30"box 4 Acer p. 'Sango Kaku' 10-gal 44 Acer p. 'Scolopendrifolium' 30"box 2 Acer p. 'Shishigashira' 24"box/25gal 3 Acer p. 'Shishigashira' multi 30"box 1 Acer p. 'Suminagashi' 25gal/24"box 4 Acer p.d. 'Crimson Queen' 15-gal 2 Acer p.d. 'Red Dragon' 15-gal 3 Acer p.d. 'Sekimori' 24"box/25gal 2 Acer p.d. 'Sekimori' 30"box 1 Acer p.d. 'Spring Delight' 24"box/25gal 2 Acer p.d. -

Variability, Heritability and Genetic Advance Studies in Single Type
International Journal of Chemical Studies 2020; 8(1): 3105-3108 P-ISSN: 2349–8528 E-ISSN: 2321–4902 www.chemijournal.com Variability, heritability and genetic advance IJCS 2020; 8(1): 3105-3108 © 2020 IJCS studies in single type genotypes of tuberose (Agave Received: 28-11-2019 Accepted: 30-12-2019 amica (Medik.) Syn. Polianthes tuberosa L.) Ratna Priyanka R Ph.D. Scholar, Department of Ratna Priyanka R, M Kannan, M Ganga and N Kumaravadivel Floriculture and Landscape Architecture, Tamil Nadu DOI: https://doi.org/10.22271/chemi.2020.v8.i1au.8743 Agricultural University, Coimbatore, Tamil Nadu, India Abstract M Kannan The present investigation on performance and genetic variability studies in tuberose (Agave amica Professor, Directorate of (Medik.) Syn. Polianthes tuberosa L.) Genotypes was conducted at Botanical Garden, Department of Research, Tamil Nadu Floriculture and Landscape Architecture, Tamil Nadu Agricultural University during the year 2018-2020. Agricultural University, Among seventeen single type genotypes, Prajwal and Bidhan Rajani-I showed better performance in Coimbatore, Tamil Nadu, India vegetative and flowering characters and recorded comparatively high loose flower yield of 1.05 and 1.00 kg/m2 respectively. Among different vegetative and flowering parameters recorded, high phenotypic M Ganga coefficient of variation (PCV) and genotypic coefficient of variation (GCV) were recorded in weight of Associate Professor (Hort.), ten florets and loose flower yield per square meter whereas moderate values were observed in spike Department of Floriculture and length, rachis length, number of florets per spike and spike weight were recorded moderate. However, in Landscape Architecture, Tamil all the parameters PCV is more than GCV.