Amino Acid Catabolism-Directed Biofuel Production in Clostridium Sticklandii: an Insight Into Model-Driven Systems Engineering
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

QM/MM) Study on Ornithine Cyclodeaminase (OCD): a Tale of Two Iminiums
Int. J. Mol. Sci. 2012, 13, 12994-13011; doi:10.3390/ijms131012994 OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Article A Molecular Dynamics (MD) and Quantum Mechanics/Molecular Mechanics (QM/MM) Study on Ornithine Cyclodeaminase (OCD): A Tale of Two Iminiums Bogdan F. Ion, Eric A. C. Bushnell, Phil De Luna and James W. Gauld * Department of Chemistry and Biochemistry, University of Windsor, Windsor, ON N9B 3P4, Canada; E-Mails: [email protected] (B.F.I.); [email protected] (E.A.C.B.); [email protected] (P.D.L.) * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +1-519-253-3000; Fax: +1-519-973-7098. Received: 10 September 2012; in revised form: 27 September 2012 / Accepted: 27 September 2012 / Published: 11 October 2012 Abstract: Ornithine cyclodeaminase (OCD) is an NAD+-dependent deaminase that is found in bacterial species such as Pseudomonas putida. Importantly, it catalyzes the direct conversion of the amino acid L-ornithine to L-proline. Using molecular dynamics (MD) and a hybrid quantum mechanics/molecular mechanics (QM/MM) method in the ONIOM formalism, the catalytic mechanism of OCD has been examined. The rate limiting step is calculated to be the initial step in the overall mechanism: hydride transfer from the + + L-ornithine’s Cα–H group to the NAD cofactor with concomitant formation of a Cα=NH2 Schiff base with a barrier of 90.6 kJ mol−1. Importantly, no water is observed within the + active site during the MD simulations suitably positioned to hydrolyze the Cα=NH2 intermediate to form the corresponding carbonyl. -

Acetyl-Coa Synthetase 3 Promotes Bladder Cancer Cell Growth Under Metabolic Stress Jianhao Zhang1, Hongjian Duan1, Zhipeng Feng1,Xinweihan1 and Chaohui Gu2
Zhang et al. Oncogenesis (2020) 9:46 https://doi.org/10.1038/s41389-020-0230-3 Oncogenesis ARTICLE Open Access Acetyl-CoA synthetase 3 promotes bladder cancer cell growth under metabolic stress Jianhao Zhang1, Hongjian Duan1, Zhipeng Feng1,XinweiHan1 and Chaohui Gu2 Abstract Cancer cells adapt to nutrient-deprived tumor microenvironment during progression via regulating the level and function of metabolic enzymes. Acetyl-coenzyme A (AcCoA) is a key metabolic intermediate that is crucial for cancer cell metabolism, especially under metabolic stress. It is of special significance to decipher the role acetyl-CoA synthetase short chain family (ACSS) in cancer cells confronting metabolic stress. Here we analyzed the generation of lipogenic AcCoA in bladder cancer cells under metabolic stress and found that in bladder urothelial carcinoma (BLCA) cells, the proportion of lipogenic AcCoA generated from glucose were largely reduced under metabolic stress. Our results revealed that ACSS3 was responsible for lipogenic AcCoA synthesis in BLCA cells under metabolic stress. Interestingly, we found that ACSS3 was required for acetate utilization and histone acetylation. Moreover, our data illustrated that ACSS3 promoted BLCA cell growth. In addition, through analyzing clinical samples, we found that both mRNA and protein levels of ACSS3 were dramatically upregulated in BLCA samples in comparison with adjacent controls and BLCA patients with lower ACSS3 expression were entitled with longer overall survival. Our data revealed an oncogenic role of ACSS3 via regulating AcCoA generation in BLCA and provided a promising target in metabolic pathway for BLCA treatment. 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; Introduction acetyl-CoA synthetase short chain family (ACSS), which In cancer cells, considerable number of metabolic ligates acetate and CoA6. -

Part One Amino Acids As Building Blocks
Part One Amino Acids as Building Blocks Amino Acids, Peptides and Proteins in Organic Chemistry. Vol.3 – Building Blocks, Catalysis and Coupling Chemistry. Edited by Andrew B. Hughes Copyright Ó 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim ISBN: 978-3-527-32102-5 j3 1 Amino Acid Biosynthesis Emily J. Parker and Andrew J. Pratt 1.1 Introduction The ribosomal synthesis of proteins utilizes a family of 20 a-amino acids that are universally coded by the translation machinery; in addition, two further a-amino acids, selenocysteine and pyrrolysine, are now believed to be incorporated into proteins via ribosomal synthesis in some organisms. More than 300 other amino acid residues have been identified in proteins, but most are of restricted distribution and produced via post-translational modification of the ubiquitous protein amino acids [1]. The ribosomally encoded a-amino acids described here ultimately derive from a-keto acids by a process corresponding to reductive amination. The most important biosynthetic distinction relates to whether appropriate carbon skeletons are pre-existing in basic metabolism or whether they have to be synthesized de novo and this division underpins the structure of this chapter. There are a small number of a-keto acids ubiquitously found in core metabolism, notably pyruvate (and a related 3-phosphoglycerate derivative from glycolysis), together with two components of the tricarboxylic acid cycle (TCA), oxaloacetate and a-ketoglutarate (a-KG). These building blocks ultimately provide the carbon skeletons for unbranched a-amino acids of three, four, and five carbons, respectively. a-Amino acids with shorter (glycine) or longer (lysine and pyrrolysine) straight chains are made by alternative pathways depending on the available raw materials. -

Synthetic Biology Applications in Industrial Microbiology
SYNTHETIC BIOLOGY APPLICATIONS IN INDUSTRIAL MICROBIOLOGY Topic Editors Weiwen Zhang and David R. Nielsen MICROBIOLOGY FRONTIERS COPYRIGHT STATEMENT ABOUT FRONTIERS © Copyright 2007-2014 Frontiers is more than just an open-access publisher of scholarly articles: it is a pioneering Frontiers Media SA. All rights reserved. approach to the world of academia, radically improving the way scholarly research is managed. All content included on this site, such as The grand vision of Frontiers is a world where all people have an equal opportunity to seek, share text, graphics, logos, button icons, images, and generate knowledge. Frontiers provides immediate and permanent online open access to all video/audio clips, downloads, data compilations and software, is the property its publications, but this alone is not enough to realize our grand goals. of or is licensed to Frontiers Media SA (“Frontiers”) or its licensees and/or subcontractors. The copyright in the text of individual articles is the property of their FRONTIERS JOURNAL SERIES respective authors, subject to a license granted to Frontiers. The Frontiers Journal Series is a multi-tier and interdisciplinary set of open-access, online The compilation of articles constituting journals, promising a paradigm shift from the current review, selection and dissemination this e-book, wherever published, as well as the compilation of all other content on processes in academic publishing. this site, is the exclusive property of All Frontiers journals are driven by researchers for researchers; therefore, they constitute a service Frontiers. For the conditions for downloading and copying of e-books from to the scholarly community. At the same time, the Frontiers Journal Series operates on a revo- Frontiers’ website, please see the Terms lutionary invention, the tiered publishing system, initially addressing specific communities of for Website Use. -

3-Iodo-Alpha-Methyl-L- Tyrosine
A Strategyfor the Study of CerebralAmino Acid Transport Using Iodine-123-Labeled Amino Acid Radiopharmaceutical: 3-Iodo-alpha-methyl-L- tyrosine Keiichi Kawai, Yasuhisa Fujibayashi, Hideo Saji, Yoshiharu Yonekura, Junji Konishi, Akiko Kubodera, and Akira Yokoyama Faculty ofPharmaceutical Sciences, Science Universityof Tokyo, Tokyo, Japan and School ofMedicine and Faculty of Pharmaceutical Sciences, Kyolo University, Kyoto, Japan In the present work, a search for radioiodinated tyrosine We examined the brain accumulation of iodine-i 23-iodo-al derivatives for the cerebral tyrosine transport is attempted. pha-methyl-L-tyrosine (123I-L-AMT)in mice and rats. l-L-AMT In the screening process, in vitro accumulation studies in showed high brain accumulation in mice, and in rats; rat brain rat brain slices, measurement ofbrain uptake index (BUI), uptake index exceeded that of 14C-L-tyrosine.The brain up and in vivo mouse biodistribution are followed by the take index and the brain slice studies indicated the affinity of analysis of metabolites. In a preliminary study, the radio l-L-AMT for earner-mediatedand stereoselective active trans iodinated monoiodotyrosine in its L- and D-form (L-MIT port systems, respectively; both operating across the blood and D-MIT) and its noniodinated counterpart (‘4C-L- brain barrier and cell membranes of the brain. The tissue tyrosine) are tested. homogenate analysis revealed that most of the accumulated radioactivity belonged to intact l-L-AMT, an indication of its The initial part of the work provided the basis for a stability. Thus, 1231-L-AMTappears to be a useful radiophar radioiodinated tyrosine derivative to be used for the meas maceutical for the selective measurement of cerebral amino urement of cerebral tyrosine transport. -

Development of a Phage Display Library for Discovery of Antigenic Brucella Peptides Jeffrey Williams Iowa State University
Iowa State University Capstones, Theses and Graduate Theses and Dissertations Dissertations 2018 Development of a phage display library for discovery of antigenic Brucella peptides Jeffrey Williams Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/etd Part of the Microbiology Commons Recommended Citation Williams, Jeffrey, "Development of a phage display library for discovery of antigenic Brucella peptides" (2018). Graduate Theses and Dissertations. 16896. https://lib.dr.iastate.edu/etd/16896 This Thesis is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Development of a phage display library for discovery of antigenic Brucella peptides by Jeffrey Williams A thesis submitted to the graduate faculty in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE Major: Microbiology Program of Study Committee: Bryan H. Bellaire, Major Professor Steven Olsen Steven Carlson The student author, whose presentation of the scholarship herein was approved by the program of study committee, is solely responsible for the content of this thesis. The Graduate College will ensure this thesis is globally accessible and will not permit alterations after a degree is conferred. Iowa State University -

New Insights Into Clostridia Through Comparative Analyses of Their 40 Genomes
Bioenerg. Res. DOI 10.1007/s12155-014-9486-9 New Insights into Clostridia Through Comparative Analyses of Their 40 Genomes Chuan Zhou & Qin Ma & Xizeng Mao & Bingqiang Liu & Yanbin Yin & Ying Xu # Springer Science+Business Media New York 2014 Abstract The Clostridium genus of bacteria contains the gene groups are shared by all the strains, and the strain- most widely studied biofuel-producing organisms such as specific gene pool consists of 22,668 genes, which is consis- Clostridium thermocellum and also some human pathogens, tent with the fact that these bacteria live in very diverse plus a few less characterized strains. Here, we present a environments and have evolved a very large number of comparative genomic analysis of 40 fully sequenced clostrid- strain-specific genes to adapt to different environments. ial genomes, paying a particular attention to the biomass Across the 40 genomes, 1.4–5.8 % of genes fall into the degradation ones. Our analysis indicates that some of the carbohydrate active enzyme (CAZyme) families, and 20 out Clostridium botulinum strains may have been incorrectly clas- of the 40 genomes may encode cellulosomes with each ge- sified in the current taxonomy and hence should be renamed nome having 1 to 76 genes bearing the cellulosome-related according to the 16S ribosomal RNA (rRNA) phylogeny. A modules such as dockerins and cohesins. A phylogenetic core-genome analysis suggests that only 169 orthologous footprinting analysis identified cis-regulatory motifs that are enriched in the promoters of the CAZyme genes, giving rise to 32 statistically significant motif candidates. Keywords Clostridium . Comparative genomics . -
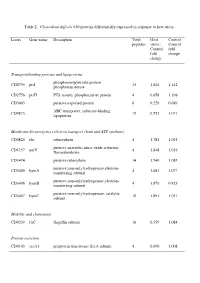
Table 2: Clostridium Difficile 630 Proteins Differentially Expressed in Response to Heat Stress
Table 2: Clostridium difficile 630 proteins differentially expressed in response to heat stress. Locus Gene name Description Total Heat Control : peptides stress : Control Control fold fold change change Transport/binding proteins and lipoproteins phosphoenolpyruvate-protein CD2755 ptsI 15 1.644 1.142 phosphotransferase CD2756 ptsH PTS system, phosphocarrier protein 4 0.658 1.198 CD3043 putative exported protein 6 0.229 0.689 ABC transporter, substrate-binding CD0873 15 0.521 1.071 lipoprotein Membrane bioenergetics (electron transport chain and ATP synthase) CD0825 rbr rubrerythrin 4 1.781 1.035 putative anaerobic nitric oxide reductase CD1157 norV 4 1.848 1.016 flavorubredoxin CD1474 putative ruberythrin 14 1.540 1.085 putative iron-only hydrogenase,electron- CD3405 hymA 4 1.681 1.077 transferring subunit putative iron-only hydrogenase,electron- CD3406 hymB 4 1.870 0.923 transferring subunit putative iron-only hydrogenase, catalytic CD3407 hymC 10 1.891 1.051 subunit Mobility and chemotaxis CD0239 fliC flagellin subunit 16 0.559 1.084 Protein secretion CD0143 secA1 preprotein translocase SecA subunit 4 0.690 1.008 Specific pathways putative oxidoreductase, thiamine diP-binding CD0116 13 1.628 1.121 subunit putative oxidoreductase, acetyl-CoA synthase CD0174 4 2.687 1.055 subunit CD0790 putative NUDIX-family hydrolase 8 1.491 1.32 inosine-uridine preferring nucleoside CD1682 iunH 4 0.518 0.984 hydrolase CD2181 putative aromatic compounds hydrolase 7 0372 0.909 CD2342 sucD succinate-semialdehyde dehydrogenase 6 0.651 0.906 Metabolism -

Gastroenterology
Gastroenterology Inhibiting Growth of Clostridioides difficile by Restoring Valerate, Produced by the Intestinal Microbiota --Manuscript Draft-- Manuscript Number: GASTRO-D-18-00449R1 Full Title: Inhibiting Growth of Clostridioides difficile by Restoring Valerate, Produced by the Intestinal Microbiota Article Type: Basic - Alimentary Tract Corresponding Author: Julian Marchesi Cardiff University Cardiff, UNITED KINGDOM Corresponding Author Secondary Information: Corresponding Author's Institution: Cardiff University Corresponding Author's Secondary Institution: First Author: Julie A. K. McDonald, PhD First Author Secondary Information: Order of Authors: Julie A. K. McDonald, PhD Benjamin H. Mullish, MB BChir Alexandros Pechlivanis, PhD Zhigang Liu, PhD Jerusa Brignardello, MSc Dina Kao, MD Elaine Holmes, PhD Jia V. Li, PhD Thomas B Clarke, PhD Mark R. Thursz, MD Julian R. Marchesi, PhD Order of Authors Secondary Information: Abstract: Background & Aims: Faecal microbiota transplantation (FMT) is effective for treating recurrent Clostridioides difficile infection (CDI), but there are concerns regarding its long-term safety. Understanding the mechanisms of the effects of FMT could help us design safer, targeted therapies. We aimed to identify microbial metabolites that are important for C difficile growth. Methods: We used a CDI chemostat model as a tool to study the effects of FMT in vitro. The following analyses were performed: C difficile plate counts, 16S rRNA gene sequencing, 1H-NMR spectroscopy, and UPLC mass spectrometry bile acid profiling. FMT mixtures were prepared using fresh faecal samples provided by donors enrolled in the FMT Programme. Results from chemostat experiments were validated using human stool samples, C difficile batch cultures, and C57BL/6 mice with CDI. Human stool samples were collected from 16 patients with recurrent CDI and 5 healthy donors participating in an FMT trial. -

Structures, Functions, and Mechanisms of Filament Forming Enzymes: a Renaissance of Enzyme Filamentation
Structures, Functions, and Mechanisms of Filament Forming Enzymes: A Renaissance of Enzyme Filamentation A Review By Chad K. Park & Nancy C. Horton Department of Molecular and Cellular Biology University of Arizona Tucson, AZ 85721 N. C. Horton ([email protected], ORCID: 0000-0003-2710-8284) C. K. Park ([email protected], ORCID: 0000-0003-1089-9091) Keywords: Enzyme, Regulation, DNA binding, Nuclease, Run-On Oligomerization, self-association 1 Abstract Filament formation by non-cytoskeletal enzymes has been known for decades, yet only relatively recently has its wide-spread role in enzyme regulation and biology come to be appreciated. This comprehensive review summarizes what is known for each enzyme confirmed to form filamentous structures in vitro, and for the many that are known only to form large self-assemblies within cells. For some enzymes, studies describing both the in vitro filamentous structures and cellular self-assembly formation are also known and described. Special attention is paid to the detailed structures of each type of enzyme filament, as well as the roles the structures play in enzyme regulation and in biology. Where it is known or hypothesized, the advantages conferred by enzyme filamentation are reviewed. Finally, the similarities, differences, and comparison to the SgrAI system are also highlighted. 2 Contents INTRODUCTION…………………………………………………………..4 STRUCTURALLY CHARACTERIZED ENZYME FILAMENTS…….5 Acetyl CoA Carboxylase (ACC)……………………………………………………………………5 Phosphofructokinase (PFK)……………………………………………………………………….6 -

Generate Metabolic Map Poster
Authors: Pallavi Subhraveti Ron Caspi Peter Midford Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_001591825Cyc: Bacillus vietnamensis NBRC 101237 Cellular Overview Connections between pathways are omitted for legibility. Anamika Kothari sn-glycerol phosphate phosphate pro phosphate phosphate phosphate thiamine molybdate D-xylose D-ribose glutathione 3-phosphate D-mannitol L-cystine L-djenkolate lanthionine α,β-trehalose phosphate phosphate [+ 3 more] α,α-trehalose predicted predicted ABC ABC FliY ThiT XylF RbsB RS10935 UgpC TreP PutP RS10200 PstB PstB RS10385 RS03335 RS20030 RS19075 transporter transporter of molybdate of phosphate α,β-trehalose 6-phosphate L-cystine D-xylose D-ribose sn-glycerol D-mannitol phosphate phosphate thiamine glutathione α α phosphate phosphate phosphate phosphate L-djenkolate 3-phosphate , -trehalose 6-phosphate pro 1-phosphate lanthionine molybdate phosphate [+ 3 more] Metabolic Regulator Amino Acid Degradation Amine and Polyamine Biosynthesis Macromolecule Modification tRNA-uridine 2-thiolation Degradation ATP biosynthesis a mature peptidoglycan a nascent β an N-terminal- -

Generated by SRI International Pathway Tools Version 25.0, Authors S
Authors: Pallavi Subhraveti Ron Caspi Quang Ong Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000725805Cyc: Streptomyces xanthophaeus Cellular Overview Connections between pathways are omitted for legibility.